Proteomic analysis of human prostate cancer PC-3M-1E8 cells and PC-3M-2B4 cells of same origin but with different metastatic potential
- PMID: 30379883
- PMCID: PMC6209233
- DOI: 10.1371/journal.pone.0206139
Proteomic analysis of human prostate cancer PC-3M-1E8 cells and PC-3M-2B4 cells of same origin but with different metastatic potential
Abstract
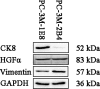
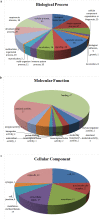
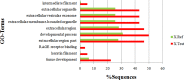
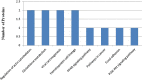
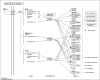
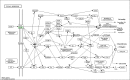
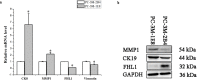
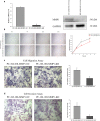
Prostate cancer (PCa) is the second most frequently diagnosed cancer and the fifth leading cause of death from cancer in men worldwide. Increased understanding of the prostate cancer metastasis mechanisms will help identify more efficient intervention strategies to prevent or treat this deadly disease in the future. To identify the candidate proteins that contribute to metastasis of PCa, isobaric tags for relative and absolute quantitation (iTRAQ)-based proteomic analysis was performed to explore differentially expressed proteins between two homologous human prostate cancer cell lines including highly-metastatic PC-3M-1E8 cell line and poorly-metastatic PC-3M-2B4 cell line. Here, a total of 58 proteins were identified to be significantly differentially expressed between PC-3M-1E8 and PC-3M-2B4 cells, which were further verified using real-time quantitative PCR and western blot analysis. The bioinformatic analysis suggested that the differentially expressed proteins, like MMP1 and FHL1, may contribute to the higher metastatic ability of PC-3M-1E8 cells than PC-3M-2B4 cells. In addition, functional analyses proved MMP1's positive effect on the higher metastatic ability of PC-3M-1E8 cells than PC-3M-2B4 cells. These findings provided a unique resource to specifically reveal the complex molecular regulatory mechanisms underlying the progression of prostate cancer from poorly-metastatic to highly-metastatic stage.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

