SymMap: an integrative database of traditional Chinese medicine enhanced by symptom mapping
- PMID: 30380087
- PMCID: PMC6323958
- DOI: 10.1093/nar/gky1021
SymMap: an integrative database of traditional Chinese medicine enhanced by symptom mapping
Abstract
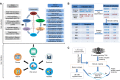
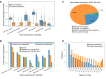
Recently, the pharmaceutical industry has heavily emphasized phenotypic drug discovery (PDD), which relies primarily on knowledge about phenotype changes associated with diseases. Traditional Chinese medicine (TCM) provides a massive amount of information on natural products and the clinical symptoms they are used to treat, which are the observable disease phenotypes that are crucial for clinical diagnosis and treatment. Curating knowledge of TCM symptoms and their relationships to herbs and diseases will provide both candidate leads and screening directions for evidence-based PDD programs. Therefore, we present SymMap, an integrative database of traditional Chinese medicine enhanced by symptom mapping. We manually curated 1717 TCM symptoms and related them to 499 herbs and 961 symptoms used in modern medicine based on a committee of 17 leading experts practicing TCM. Next, we collected 5235 diseases associated with these symptoms, 19 595 herbal constituents (ingredients) and 4302 target genes, and built a large heterogeneous network containing all of these components. Thus, SymMap integrates TCM with modern medicine in common aspects at both the phenotypic and molecular levels. Furthermore, we inferred all pairwise relationships among SymMap components using statistical tests to give pharmaceutical scientists the ability to rank and filter promising results to guide drug discovery. The SymMap database can be accessed at http://www.symmap.org/ and https://www.bioinfo.org/symmap.
Figures

References
-
- Swinney D.C. Phenotypic vs. target-based drug discovery for first-in-class medicines. Clin. Pharmacol. Ther. 2013; 93:299–301. - PubMed
-
- Swinney D.C., Anthony J.. How were new medicines discovered. Nat. Rev. Drug Discov. 2011; 10:507–519. - PubMed
-
- Harvey A.L., Edrada-Ebel R., Quinn R.J.. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015; 14:111–129. - PubMed
-
- Qiu J. Traditional medicine: a culture in the balance. Nature. 2007; 448:126–128. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

