Large Differences in Small RNA Composition Between Human Biofluids
- PMID: 30380423
- PMCID: PMC6261476
- DOI: 10.1016/j.celrep.2018.10.014
Large Differences in Small RNA Composition Between Human Biofluids
Abstract
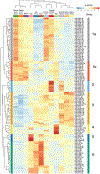
Extracellular microRNAs (miRNAs) and other small RNAs are implicated in cellular communication and may be useful as disease biomarkers. We systematically compared small RNAs in 12 human biofluid types using RNA sequencing (RNA-seq). miRNAs and tRNA-derived RNAs (tDRs) accounted for the majority of mapped reads in all biofluids, but the ratio of miRNA to tDR reads varied from 72 in plasma to 0.004 in bile. miRNA levels were highly correlated across all biofluids, but levels of some miRNAs differed markedly between biofluids. tDR populations differed extensively between biofluids. Y RNA fragments were seen in all biofluids and accounted for >10% of reads in blood plasma, serum, and cerebrospinal fluid (CSF). Reads mapping exclusively to Piwi-interacting RNAs (piRNAs) were very rare, except in seminal plasma. These results demonstrate extensive differences in small RNAs between human biofluids and provide a useful resource for investigating extracellular RNA biology and developing biomarkers.
Keywords: Y RNA; biofluids; extracellular RNA; miRNA; tRNA.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




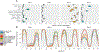
References
-
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, et al. (2011). Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 108, 5003–5008. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

