Enzymes Catalyzing the TCA- and Urea Cycle Influence the Matrix Composition of Biofilms Formed by Methicillin-Resistant Staphylococcus aureus USA300
- PMID: 30380651
- PMCID: PMC6313315
- DOI: 10.3390/microorganisms6040113
Enzymes Catalyzing the TCA- and Urea Cycle Influence the Matrix Composition of Biofilms Formed by Methicillin-Resistant Staphylococcus aureus USA300
Abstract
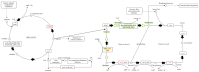
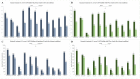
In methicillin-sensitive Staphylococcus aureus (MSSA), the tricarboxylic acid (TCA) cycle is known to negatively regulate production of the major biofilm-matrix exopolysaccharide, PIA/PNAG. However, methicillin-resistant S. aureus (MRSA) produce a primarily proteinaceous biofilm matrix, and contribution of the TCA-cycle therein remains unclear. Utilizing USA300-JE2 Tn-mutants (NARSA) in genes encoding TCA- and urea cycle enzymes for transduction into a prolific biofilm-forming USA300 strain (UAS391-Erys), we studied the contribution of the TCA- and urea cycle and of proteins, eDNA and PIA/PNAG, to the matrix. Genes targeted in the urea cycle encoded argininosuccinate lyase and arginase (argH::Tn and rocF::Tn), and in the TCA-cycle encoded succinyl-CoA synthetase, succinate dehydrogenase, aconitase, isocitrate dehydrogenase, fumarate hydratase class II, and citrate synthase II (sucC::Tn, sdhA/B::Tn, acnA::Tn, icd::Tn, fumC::Tn and gltA::Tn). Biofilm formation was significantly decreased under no flow and flow conditions by argH::Tn, fumC::Tn, and sdhA/B::Tn (range OD492 0.374-0.667; integrated densities 2.065-4.875) compared to UAS391-EryS (OD492 0.814; integrated density 10.676) (p ≤ 0.008). Cellular and matrix stains, enzymatic treatment (Proteinase K, DNase I), and reverse-transcriptase PCR-based gene-expression analysis of fibronectin-binding proteins (fnbA/B) and the staphylococcal accessory regulator (sarA) on pre-formed UAS391-Erys and Tn-mutant biofilms showed: (i) < 1% PIA/PNAG in the proteinaceous/eDNA matrix; (ii) increased proteins under no flow and flow in the matrix of Tn mutant biofilms (on average 50 and 51 (±11)%) compared to UAS391-Erys (on average 22 and 25 (±4)%) (p < 0.001); and (iii) down- and up-regulation of fnbA/B and sarA, respectively, in Tn-mutants compared to UAS391-EryS (0.62-, 0.57-, and 2.23-fold on average). In conclusion, we show that the biofilm matrix of MRSA-USA300 and the corresponding Tn mutants is PIA/PNAG-independent and are mainly composed of proteins and eDNA. The primary impact of TCA-cycle inactivation was on the protein component of the biofilm matrix of MRSA-USA300.
Keywords: MRSA; MSSA; SCCmec; argH; arginine; fnbA; fnbB; fumC; fumarate; malate; sarA; sdhA; sdhB; tricarboxylic acid cycle.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Fatty acid kinase A is an important determinant of biofilm formation in Staphylococcus aureus USA300.BMC Genomics. 2015 Oct 26;16:861. doi: 10.1186/s12864-015-1956-8. BMC Genomics. 2015. PMID: 26502874 Free PMC article.
-
Genetic evidence for an alternative citrate-dependent biofilm formation pathway in Staphylococcus aureus that is dependent on fibronectin binding proteins and the GraRS two-component regulatory system.Infect Immun. 2008 Jun;76(6):2469-77. doi: 10.1128/IAI.01370-07. Epub 2008 Mar 17. Infect Immun. 2008. PMID: 18347039 Free PMC article.
-
The small non-coding RNA RsaE influences extracellular matrix composition in Staphylococcus epidermidis biofilm communities.PLoS Pathog. 2019 Mar 14;15(3):e1007618. doi: 10.1371/journal.ppat.1007618. eCollection 2019 Mar. PLoS Pathog. 2019. PMID: 30870530 Free PMC article.
-
Methicillin resistance and the biofilm phenotype in Staphylococcus aureus.Front Cell Infect Microbiol. 2015 Jan 28;5:1. doi: 10.3389/fcimb.2015.00001. eCollection 2015. Front Cell Infect Microbiol. 2015. PMID: 25674541 Free PMC article. Review.
-
Differences in Biofilm Formation by Methicillin-Resistant and Methicillin-Susceptible Staphylococcus aureus Strains.Diseases. 2023 Nov 5;11(4):160. doi: 10.3390/diseases11040160. Diseases. 2023. PMID: 37987271 Free PMC article. Review.
Cited by
-
The Antimicrobial Peptide AMP-17 Derived from Musca domestica Inhibits Biofilm Formation and Eradicates Mature Biofilm in Candida albicans.Antibiotics (Basel). 2022 Oct 25;11(11):1474. doi: 10.3390/antibiotics11111474. Antibiotics (Basel). 2022. PMID: 36358129 Free PMC article.
-
Mechanisms underlying the virulence regulation of new Vibrio alginolyticus ncRNA Vvrr1 with a comparative proteomic analysis.Emerg Microbes Infect. 2019;8(1):1604-1618. doi: 10.1080/22221751.2019.1687261. Emerg Microbes Infect. 2019. PMID: 31711375 Free PMC article.
-
Systematic review and meta-analysis of the association between biofilm formation and antibiotic resistance in MRSE Isolated from Iranian patients.Caspian J Intern Med. 2025 Mar 11;16(2):225-232. doi: 10.22088/cjim.16.2.225. eCollection 2025 Spring. Caspian J Intern Med. 2025. PMID: 40575738 Free PMC article.
-
Regulation of airway fumarate by host and pathogen promotes Staphylococcus aureus pneumonia.Nat Commun. 2025 Aug 1;16(1):7050. doi: 10.1038/s41467-025-62453-y. Nat Commun. 2025. PMID: 40745169 Free PMC article.
-
Synchronous bacterial barrier and exudate absorption: A novel dual-function dressing strategy for pin-site infection prevention.Mater Today Bio. 2025 May 8;32:101833. doi: 10.1016/j.mtbio.2025.101833. eCollection 2025 Jun. Mater Today Bio. 2025. PMID: 40487154 Free PMC article.
References
-
- O’Neill E., Pozzi C., Houston P., Smyth D., Humphreys H., Robinson D.A., O’Gara J.P. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J. Clin. Microbiol. 2007;45:1379–1388. doi: 10.1128/JCM.02280-06. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

