Comparative Evaluation of Machine Learning Strategies for Analyzing Big Data in Psychiatry
- PMID: 30380679
- PMCID: PMC6274760
- DOI: 10.3390/ijms19113387
Comparative Evaluation of Machine Learning Strategies for Analyzing Big Data in Psychiatry
Abstract
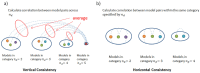
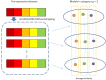
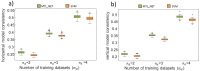
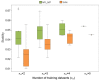
The requirement of innovative big data analytics has become a critical success factor for research in biological psychiatry. Integrative analyses across distributed data resources are considered essential for untangling the biological complexity of mental illnesses. However, little is known about algorithm properties for such integrative machine learning. Here, we performed a comparative analysis of eight machine learning algorithms for identification of reproducible biological fingerprints across data sources, using five transcriptome-wide expression datasets of schizophrenia patients and controls as a use case. We found that multi-task learning (MTL) with network structure (MTL_NET) showed superior accuracy compared to other MTL formulations as well as single task learning, and tied performance with support vector machines (SVM). Compared to SVM, MTL_NET showed significant benefits regarding the variability of accuracy estimates, as well as its robustness to cross-dataset and sampling variability. These results support the utility of this algorithm as a flexible tool for integrative machine learning in psychiatry.
Keywords: biomarker discovery; machine learning; multi-task learning; psychiatry.
Conflict of interest statement
A.M.-L. has received consultant fees from Blueprint Partnership, Boehringer Ingelheim, Daimler und Benz Stiftung, Elsevier, F. Hoffmann-La Roche, ICARE Schizophrenia, K. G. Jebsen Foundation, L.E.K Consulting, Lundbeck International Foundation (LINF), R. Adamczak, Roche Pharma, Science Foundation, Synapsis Foundation—Alzheimer Research Switzerland, System Analytics, and has received lectures including travel fees from Boehringer Ingelheim, Fama Public Relations, Institut d'investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), Janssen-Cilag, Klinikum Christophsbad, Göppingen, Lilly Deutschland, Luzerner Psychiatrie, LVR Klinikum Düsseldorf, LWL PsychiatrieVerbund Westfalen-Lippe, Otsuka Pharmaceuticals, Reunions i Ciencia S. L., Spanish Society of Psychiatry, Südwestrundfunk Fernsehen, Stern TV, and Vitos Klinikum Kurhessen. All other authors declare no potential conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures



References
-
- Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium. Ripke S., Wray N.R., Lewis C.M., Hamilton S.P., Weissman M.M., Breen G., Byrne E.M., Blackwood D.H., Boomsma D.I., et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry. 2013;18:497–511. - PMC - PubMed
-
- Wolfers T., Buitelaar J.K., Beckmann C.F., Franke B., Marquand A.F. From estimating activation locality to predicting disorder: A review of pattern recognition for neuroimaging-based psychiatric diagnostics. Neurosci. Biobehav. Rev. 2015;57:328–349. doi: 10.1016/j.neubiorev.2015.08.001. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

