Identification and Expression Analyses of SBP-Box Genes Reveal Their Involvement in Abiotic Stress and Hormone Response in Tea Plant (Camellia sinensis)
- PMID: 30380795
- PMCID: PMC6274802
- DOI: 10.3390/ijms19113404
Identification and Expression Analyses of SBP-Box Genes Reveal Their Involvement in Abiotic Stress and Hormone Response in Tea Plant (Camellia sinensis)
Abstract
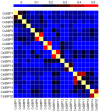
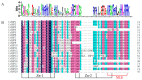
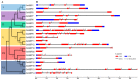
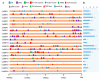
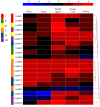
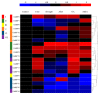
The SQUAMOSA promoter binding protein (SBP)-box gene family is a plant-specific transcription factor family. This family plays a crucial role in plant growth and development. In this study, 20 SBP-box genes were identified in the tea plant genome and classified into six groups. The genes in each group shared similar exon-intron structures and motif positions. Expression pattern analyses in five different tissues demonstrated that expression in the buds and leaves was higher than that in other tissues. The cis-elements and expression patterns of the CsSBP genes suggested that the CsSBP genes play active roles in abiotic stress responses; these responses may depend on the abscisic acid (ABA), gibberellic acid (GA), and methyl jasmonate (MeJA) signaling pathways. Our work provides a comprehensive understanding of the CsSBP family and will aid in genetically improving tea plants.
Keywords: Camellia sinensis; SBP-box; abiotic stress; expression profile; hormone.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

