A Nonribosomal Peptide Synthase Gene Driving Virulence in Mycobacterium tuberculosis
- PMID: 30381350
- PMCID: PMC6211224
- DOI: 10.1128/mSphere.00352-18
A Nonribosomal Peptide Synthase Gene Driving Virulence in Mycobacterium tuberculosis
Abstract
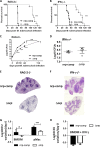
Nonribosomal peptide synthases produce short peptides in a manner that is distinct from classical mRNA-dependent ribosome-mediated translation. The Mycobacterium tuberculosis genome harbors a nonribosomal peptide synthase gene, nrp, which is part of a gene cluster proposed to be involved in the biosynthesis of isonitrile lipopeptides. Orthologous clusters are found in other slow-growing pathogenic mycobacteria and actinomycetes. To probe the role of the nrp gene in infection, we generated an nrp deletion mutant in M. tuberculosis H37Rv and tested its virulence in immunocompetent (C57BL/6) mice. The nrp mutant strain displayed lower initial growth rates in the lungs and a defective dissemination to the spleens of infected mice. Mice infected with the mutant strain also survived for twice as long as those infected with wild-type M. tuberculosis and, remarkably, showed subdued pathology, despite similar bacterial loads at later stages of infection. The differences in the course of infection between wild-type and nrp mutant strains were accompanied by distinct dynamics of the immune response. Most strikingly, the nrp mutant was highly attenuated in immunodeficient (SCID-, recombination activating 2 [RAG2]-, and gamma interferon [IFN-γ]-deficient) mice, suggesting that macrophages control the nrp mutant more efficiently than they control the wild-type strain. However, in the presence of IFN-γ, both strains were equally controlled. We propose that the nrp gene and its associated cluster are drivers of virulence during the early stages of infection.IMPORTANCE Over 10 million people developed tuberculosis (TB) in 2016, and over 1.8 million individuals succumbed to the disease. These numbers make TB the ninth leading cause of death worldwide and the leading cause from a single infectious agent. Therefore, finding novel therapeutic targets in Mycobacterium tuberculosis, the pathogen that causes most cases of human TB, is critical. In this study, we reveal a novel virulence factor in M. tuberculosis, the nrp gene. The lack of nrp highly attenuates the course of M. tuberculosis infection in the mouse model, which is particularly relevant in immune-deficient hosts. This is very relevant as TB is particularly incident in immune-suppressed individuals, such as HIV patients.
Keywords: immune deficiency; pathogenesis; tuberculosis; virulence factors.
Copyright © 2018 Bhatt et al.
Figures

References
-
- World Health Organization. 2017. Global tuberculosis control. World Health Organization, Geneva, Switzerland.
-
- DeJesus MA, Gerrick ER, Xu W, Park SW, Long JE, Boutte CC, Rubin EJ, Schnappinger D, Ehrt S, Fortune SM, Sassetti CM, Ioerger TR. 2017. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 8:e02133-16. doi:10.1128/mBio.02133-16. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

