Profilin binding couples chloride intracellular channel protein CLIC4 to RhoA-mDia2 signaling and filopodium formation
- PMID: 30381396
- PMCID: PMC6302171
- DOI: 10.1074/jbc.RA118.002779
Profilin binding couples chloride intracellular channel protein CLIC4 to RhoA-mDia2 signaling and filopodium formation
Abstract
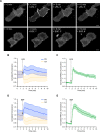
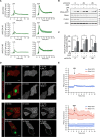
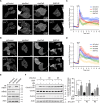
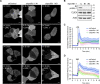
Chloride intracellular channel 4 (CLIC4) is a cytosolic protein implicated in diverse actin-based processes, including integrin trafficking, cell adhesion, and tubulogenesis. CLIC4 is rapidly recruited to the plasma membrane by RhoA-activating agonists and then partly colocalizes with β1 integrins. Agonist-induced CLIC4 translocation depends on actin polymerization and requires conserved residues that make up a putative binding groove. However, the mechanism and significance of CLIC4 trafficking have been elusive. Here, we show that RhoA activation by either lysophosphatidic acid (LPA) or epidermal growth factor is necessary and sufficient for CLIC4 translocation to the plasma membrane and involves regulation by the RhoA effector mDia2, a driver of actin polymerization and filopodium formation. We found that CLIC4 binds the G-actin-binding protein profilin-1 via the same residues that are required for CLIC4 trafficking. Consistently, shRNA-induced profilin-1 silencing impaired agonist-induced CLIC4 trafficking and the formation of mDia2-dependent filopodia. Conversely, CLIC4 knockdown increased filopodium formation in an integrin-dependent manner, a phenotype rescued by wild-type CLIC4 but not by the trafficking-incompetent mutant CLIC4(C35A). Furthermore, CLIC4 accelerated LPA-induced filopodium retraction. We conclude that through profilin-1 binding, CLIC4 functions in a RhoA-mDia2-regulated signaling network to integrate cortical actin assembly and membrane protrusion. We propose that agonist-induced CLIC4 translocation provides a feedback mechanism that counteracts formin-driven filopodium formation.
Keywords: G protein-coupled receptor; G-actin; Rho (Rho GTPase); actin; cell adhesion; cell motility; epidermal growth factor (EGF); filopodia; formin; lysophosphatidic acid; membrane trafficking; profilin.
© 2018 Argenzio et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures




References
-
- Harrop S. J., DeMaere M. Z., Fairlie W. D., Reztsova T., Valenzuela S. M., Mazzanti M., Tonini R., Qiu M. R., Jankova L., Warton K., Bauskin A. R., Wu W. M., Pankhurst S., Campbell T. J., Breit S. N., and Curmi P. M. (2001) Crystal structure of a soluble form of the intracellular chloride ion channel CLIC1 (NCC27) at 1.4-A resolution. J. Biol. Chem. 276, 44993–45000 10.1074/jbc.M107804200 - DOI - PubMed
-
- Littler D. R., Harrop S. J., Goodchild S. C., Phang J. M., Mynott A. V., Jiang L., Valenzuela S. M., Mazzanti M., Brown L. J., Breit S. N., and Curmi P. M. (2010) The enigma of the CLIC proteins: ion channels, redox proteins, enzymes, scaffolding proteins? FEBS Lett. 584, 2093–2101 10.1016/j.febslet.2010.01.027 - DOI - PubMed
-
- Jiang L., Phang J. M., Yu J., Harrop S. J., Sokolova A. V., Duff A. P., Wilk K. E., Alkhamici H., Breit S. N., Valenzuela S. M., Brown L. J., and Curmi P. M. (2014) CLIC proteins, ezrin, radixin, moesin and the coupling of membranes to the actin cytoskeleton: a smoking gun? Biochim. Biophys. Acta 1838, 643–657 10.1016/j.bbamem.2013.05.025 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

