Metagenomic Sequencing of HIV-1 in the Blood and Female Genital Tract Reveals Little Quasispecies Diversity during Acute Infection
- PMID: 30381486
- PMCID: PMC6321908
- DOI: 10.1128/JVI.00804-18
Metagenomic Sequencing of HIV-1 in the Blood and Female Genital Tract Reveals Little Quasispecies Diversity during Acute Infection
Abstract
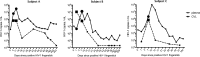
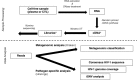
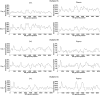
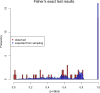
Heterosexual transmission of human immunodeficiency virus type 1 (HIV-1) is associated with a significant bottleneck in the viral quasispecies population, yet the timing of that bottleneck is poorly understood. We characterized HIV-1 diversity in the blood and female genital tract (FGT) within 2 weeks after detection of infection in three women enrolled in a unique prospective cohort in South Africa. We assembled full-length HIV-1 genomes from matched cervicovaginal lavage (CVL) samples and plasma. Deep sequencing allowed us to identify intrahost single-nucleotide variants (iSNVs) and to characterize within-sample HIV-1 diversity. Our results demonstrated very little HIV-1 diversity in the FGT and plasma by the time viremia was detectable. Within each subject, the consensus HIV-1 sequences were identical in plasma and CVL fluid. No iSNV was present at >6% frequency. One subject had 77 low-frequency iSNVs across both CVL fluid and plasma, another subject had 14 iSNVs in only CVL fluid from the earliest time point, and the third subject had no iSNVs in CVL fluid or plasma. Overall, the small amount of diversity that we detected was greater in the FGT than in plasma and declined over the first 2 weeks after viremia was detectable, compatible with a very early HIV-1 transmission bottleneck. To our knowledge, our study represents the earliest genomic analysis of HIV-1 in the FGT after transmission. Further, the use of metagenomic sequencing allowed us to characterize other organisms in the FGT, including commensal bacteria and sexually transmitted infections, highlighting the utility of the method to sequence both HIV-1 and its metagenomic environment.IMPORTANCE Due to error-prone replication, HIV-1 generates a diverse population of viruses within a chronically infected individual. When HIV-1 is transmitted to a new individual, one or a few viruses establish the new infection, leading to a genetic bottleneck in the virus population. Understanding the timing and nature of this bottleneck may provide insight into HIV-1 vaccine design and other preventative strategies. We examined the HIV-1 population in three women enrolled in a unique prospective cohort in South Africa who were followed closely during the earliest stages of HIV-1 infection. We found very little HIV-1 diversity in the blood and female genital tract during the first 2 weeks after virus was detected in the bloodstream. These results are compatible with a very early HIV-1 population bottleneck, suggesting the need to study the HIV-1 population in the female genital tract before virus is detectable in the bloodstream.
Keywords: bottleneck; female genital tract; human immunodeficiency virus; metagenomic.
Copyright © 2019 Piantadosi et al.
Figures




References
-
- Carlson JM, Schaefer M, Monaco DC, Batorsky R, Claiborne DT, Prince J, Deymier MJ, Ende ZS, Klatt NR, DeZiel CE, Lin T-H, Peng J, Seese AM, Shapiro R, Frater J, Ndung’u T, Tang J, Goepfert P, Gilmour J, Price MA, Kilembe W, Heckerman D, Goulder PJR, Allen TM, Allen S, Hunter E. 2014. HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science 345:1254031. doi:10.1126/science.1254031. - DOI - PMC - PubMed
-
- Tully DC, Ogilvie CB, Batorsky RE, Bean DJ, Power KA, Ghebremichael M, Bedard HE, Gladden AD, Seese AM, Amero MA, Lane K, McGrath G, Bazner SB, Tinsley J, Lennon NJ, Henn MR, Brumme ZL, Norris PJ, Rosenberg ES, Mayer KH, Jessen H, Kosakovsky Pond SL, Walker BD, Altfeld M, Carlson JM, Allen TM. 2016. Differences in the selection bottleneck between modes of sexual transmission influence the genetic composition of the HIV-1 founder virus. PLoS Pathog 12:e1005619. doi:10.1371/journal.ppat.1005619. - DOI - PMC - PubMed
-
- Deymier MJ, Ende Z, Fenton-May AE, Dilernia DA, Kilembe W, Allen SA, Borrow P, Hunter E. 2015. Heterosexual transmission of subtype C HIV-1 selects consensus-like variants without increased replicative capacity or interferon-α resistance. PLoS Pathog 11:e1005154. doi:10.1371/journal.ppat.1005154. - DOI - PMC - PubMed
-
- Janes H, Herbeck JT, Tovanabutra S, Thomas R, Frahm N, Duerr A, Hural J, Corey L, Self SG, Buchbinder SP, McElrath MJ, O'Connell RJ, Paris RM, Rerks-Ngarm S, Nitayaphan S, Pitisuttihum P, Kaewkungwal J, Robb ML, Michael NL, Mullins JI, Kim JH, Gilbert PB, Rolland M. 2015. HIV-1 infections with multiple founders are associated with higher viral loads than infections with single founders. Nat Med 21:1139–1141. doi:10.1038/nm.3932. - DOI - PMC - PubMed
-
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping L-H, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 105:7552–7557. doi:10.1073/pnas.0802203105. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

