A potential role of extended simple sequence repeats in competing endogenous RNA crosstalk
- PMID: 30381983
- PMCID: PMC6284579
- DOI: 10.1080/15476286.2018.1536593
A potential role of extended simple sequence repeats in competing endogenous RNA crosstalk
Abstract
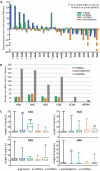
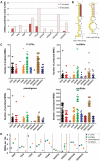
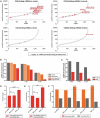
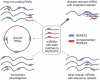
MicroRNA (miRNA)-mediated crosstalk between coding and non-coding RNAs of various types is known as the competing endogenous RNA (ceRNA) concept. Here, we propose that there is a specific variant of the ceRNA language that takes advantage of simple sequence repeat (SSR) wording. We applied bioinformatics tools to identify human transcripts that may be regarded as repeat-associated ceRNAs (raceRNAs). Multiple protein-coding transcripts, transcribed pseudogenes, long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) showing this potential were identified, and numerous miRNAs were predicted to bind to SSRs. We propose that simple repeats expanded in various hereditary neurological diseases may act as sponges for miRNAs containing complementary repeats that would affect raceRNA crosstalk. Based on the representation of specific SSRs in transcripts, expression data for SSR-binding miRNAs and expression profiling data from patients, we determined that raceRNA crosstalk is most likely to be perturbed in the case of myotonic dystrophy type 1 (DM1) and type 2 (DM2).
Keywords: ceRNA hypothesis; miRNA cooperativity; miRNA sponge; microsatellite repeats; myotonic dystrophy; non-coding RNAs; repeat expansion diseases.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
