Mating-type switching by homology-directed recombinational repair: a matter of choice
- PMID: 30382337
- PMCID: PMC6420890
- DOI: 10.1007/s00294-018-0900-2
Mating-type switching by homology-directed recombinational repair: a matter of choice
Abstract
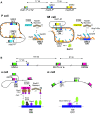
In eukaryotes, all DNA transactions happen in the context of chromatin that often takes part in regulatory mechanisms. In particular, chromatin structure can regulate exchanges of DNA occurring through homologous recombination. Few systems have provided as detailed a view on this phenomenon as mating-type switching in yeast. Mating-type switching entails the choice of a template for the gene conversions of the expressed mating-type locus. In the fission yeast Schizosaccharomyces pombe, correct template choice requires two competing small recombination enhancers, SRE2 and SRE3, that function in the context of heterochromatin. These two enhancers act with the Swi2/Swi5 recombination accessory complex to initiate strand exchange in a cell-type-specific manner, from SRE2 in M cells and SRE3 in P cells. New research indicates that the Set1C complex, responsible for H3K4 methylation, and the Brl2 ubiquitin ligase, that catalyzes H2BK119 ubiquitylation, participate in the cell-type-specific selection of SRE2 or SRE3. Here, we review these findings, compare donor preference in S. pombe to the distantly related budding yeast Saccharomyces cerevisiae, and contrast the positive effects of heterochromatin on the donor selection process with other situations, where heterochromatin represses recombination.
Keywords: Chromatin structure; Gene conversion; Histone modifications; Homology-directed repair; Mating-type switching; Recombination.
Figures


References
-
- Aboussekhra A, Chanet R, Adjiri A, Fabre F. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol Cell Biol. 1992;12:3224–3234. doi: 10.1128/MCB.12.7.3224. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

