Microglia Induce PDGFRB Expression in Glioma Cells to Enhance Their Migratory Capacity
- PMID: 30384135
- PMCID: PMC6214839
- DOI: 10.1016/j.isci.2018.10.011
Microglia Induce PDGFRB Expression in Glioma Cells to Enhance Their Migratory Capacity
Abstract
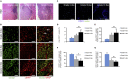
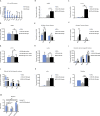
High-grade gliomas (HGGs) are the most aggressive and invasive primary brain tumors. The platelet-derived growth factor (PDGF) signaling pathway drives HGG progression, and enhanced expression of PDGF receptors (PDGFRs) is a well-established aberration in a subset of glioblastomas (GBMs). PDGFRA is expressed in glioma cells, whereas PDGFRB is mostly restricted to the glioma-associated stroma. Here we show that the spatial location of TAMMs correlates with the expansion of a subset of tumor cells that have acquired expression of PDGFRB in both mouse and human low-grade glioma and HCGs. Furthermore, M2-polarized microglia but not bone marrow (BM)-derived macrophages (BMDMs) induced PDGFRB expression in glioma cells and stimulated their migratory capacity. These findings illustrate a heterotypic cross-talk between microglia and glioma cells that may enhance the migratory and invasive capacity of the latter by inducing PDGFRB.
Keywords: Cancer; Immunology; Molecular Mechanism of Behavior; Pathophysiology.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures





Similar articles
-
Platelet-Derived Growth Factor Receptor and Ionizing Radiation in High Grade Glioma Cell Lines.Int J Mol Sci. 2019 Sep 20;20(19):4663. doi: 10.3390/ijms20194663. Int J Mol Sci. 2019. PMID: 31547056 Free PMC article.
-
Platelet-derived growth factor receptor-beta is induced during tumor development and upregulated during tumor progression in endothelial cells in human gliomas.Lab Invest. 1992 Oct;67(4):529-34. Lab Invest. 1992. PMID: 1434531
-
Pdgfra and Pdgfrb genetically interact during craniofacial development.Dev Dyn. 2016 Jun;245(6):641-52. doi: 10.1002/dvdy.24403. Epub 2016 Apr 5. Dev Dyn. 2016. PMID: 26971580 Free PMC article.
-
Peritumoral Microenvironment in High-Grade Gliomas: From FLAIRectomy to Microglia-Glioma Cross-Talk.Brain Sci. 2021 Feb 6;11(2):200. doi: 10.3390/brainsci11020200. Brain Sci. 2021. PMID: 33561993 Free PMC article. Review.
-
Platelet-derived growth factor in human glioma.Glia. 1995 Nov;15(3):257-63. doi: 10.1002/glia.440150307. Glia. 1995. PMID: 8586462 Review.
Cited by
-
A Prediction Model for Deciphering Intratumoral Heterogeneity Derived from the Microglia/Macrophages of Glioma Using Non-Invasive Radiogenomics.Brain Sci. 2023 Dec 1;13(12):1667. doi: 10.3390/brainsci13121667. Brain Sci. 2023. PMID: 38137116 Free PMC article.
-
Phase I study of vinblastine in combination with nilotinib in children, adolescents, and young adults with refractory or recurrent low-grade glioma.Neurooncol Adv. 2020 Jun 9;2(1):vdaa075. doi: 10.1093/noajnl/vdaa075. eCollection 2020 Jan-Dec. Neurooncol Adv. 2020. PMID: 32666050 Free PMC article.
-
From protein-protein interactions to immune modulation: Therapeutic prospects of targeting Neuropilin-1 in high-grade glioma.Front Immunol. 2022 Sep 20;13:958620. doi: 10.3389/fimmu.2022.958620. eCollection 2022. Front Immunol. 2022. PMID: 36203599 Free PMC article. Review.
-
Context-Dependent Glioblastoma-Macrophage/Microglia Symbiosis and Associated Mechanisms.Trends Immunol. 2021 Apr;42(4):280-292. doi: 10.1016/j.it.2021.02.004. Epub 2021 Mar 1. Trends Immunol. 2021. PMID: 33663953 Free PMC article. Review.
-
Piezo1-related physiological and pathological processes in glioblastoma.Front Cell Dev Biol. 2025 Feb 21;13:1536320. doi: 10.3389/fcell.2025.1536320. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40061015 Free PMC article. Review.
References
-
- Ajami B., Bennett J.L., Krieger C., Tetzlaff W., Rossi F.M. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci. 2007;10:1538–1543. - PubMed
-
- Burton J.L., Madsen S.A., Yao J., Sipkovsky S.S., Coussens P.M. An immunogenomics approach to understanding periparturient immunosuppression and mastitis susceptibility in dairy cows. Acta Vet. Scand. 2001;42:407–424. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

