Population genomics and climate adaptation of a C4 perennial grass, Panicum hallii (Poaceae)
- PMID: 30384830
- PMCID: PMC6211516
- DOI: 10.1186/s12864-018-5179-7
Population genomics and climate adaptation of a C4 perennial grass, Panicum hallii (Poaceae)
Abstract
Background: Understanding how and why genetic variation is partitioned across geographic space is of fundamental importance to understanding the nature of biological species. How geographical isolation and local adaptation contribute to the formation of ecotypically differentiated groups of plants is just beginning to be understood through population genomic studies. We used whole genome sequencing combined with association study of climate to discover the drivers of differentiation in the perennial C4 grass Panicum hallii.
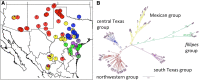
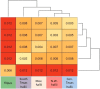
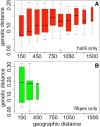
Results: Sequencing of 89 natural accessions of P.hallii revealed complex population structure across the species range. Major population genomic separation was found between subspecies P.hallii var. hallii and var. filipes as well as between at least four major unrecognized subgroups within var. hallii. At least 139 genomic SNPs were significantly associated with temperature or precipitation across the range and these SNPs were enriched for non-synonymous substitutions. SNPs associated with temperature and aridity were more often found in or near genes than expected by chance and enriched for putative involvement in dormancy processes, seed maturation, response to hyperosmosis and salinity, abscisic acid metabolism, hormone metabolism, and drought recovery.
Conclusions: Both geography and climate adaptation contribute significantly to patterns of genome-wide variation in P.hallii. Population subgroups within P.hallii may represent early stages in the formation of ecotypes. Climate associated loci identified here represent promising targets for future research in this and other perennial grasses.
Keywords: Climate GWAS; Ecotypes; Grasses; Local adaptation; Population genomics.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
-
- Turesson G. The genotypic response of the plant species to habitat. Hereditas. 1922;3:211–350. doi: 10.1111/j.1601-5223.1922.tb02734.x. - DOI
-
- Clausen J. Stages in the evolution of plant species. Ithaca: Cornell University Press; 1951.
-
- Wadgymar S, Lowry DB, Gould BA, Byron CN, Mactavish R, Anderson JT. Identifying targets and agents of selection: innovative methods to evaluate the environmental and genetic factors that contribute to local adaptation. Methods Ecol Evol. 2017;8:738–749. doi: 10.1111/2041-210X.12777. - DOI
-
- Linhart YB, Grant MC. Evolutionary significance of local genetic differentiation in plants. Annu Rev Ecol Syst. 1996;27:237–277. doi: 10.1146/annurev.ecolsys.27.1.237. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

