Identification of gammaherpesvirus infection in free-ranging black bears (Ursus americanus)
- PMID: 30385363
- PMCID: PMC7114836
- DOI: 10.1016/j.virusres.2018.10.016
Identification of gammaherpesvirus infection in free-ranging black bears (Ursus americanus)
Abstract
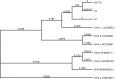
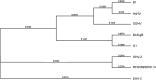
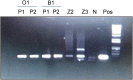
Herpesvirus infection was investigated in black bears (Ursus americanus) with neurological signs and brain lesions of nonsuppurative encephalitis of unknown cause. Visible cytopathic effects (CPE) could only be observed on days 3-5 post-infection in HrT-18G cell line inoculated with bear tissue extracts. The observed CPE in HrT-18G cells included syncytia, intranuclear inclusions, and cell detachments seen in herpesvirus infection in vitro. Herpesvirus-like particles were observed in viral culture supernatant under the electron microscope, however, capsids ranging from 60 nm to 100 nm in size were often observed in viral cultures within the first two passages of propagation. Herpesvirus infection in the bear tissues and tissue cultures were detected by PCR using degenerate primers specific to the DNA polymerase gene (DPOL) and glycoprotein B gene (gB). DNA sequencing of the amplicon revealed that the detected herpesvirus has 94-95% identity to Ursid gammaherpesvirus 1 (UrHV-1) DNA sequences of DPOL. Phylogenetic analysis of DPOL sequences indicates that black bear herpesviruses and UrHV-1 are closely related and have small distances to members of Rhadinovirus. Interestingly, black bear herpesvirus infections were also found in bears without neurological signs. The DPOL DNA sequence of black bear herpesviruses detected in neurological bears were similar to the those detected in the non-neurological bears. However, the gB DNA sequence detected from the neurological bear is different from non-neurological bear and has only 64.5%-70% identity to each other. It is possible that at least two different types of gammaherpesviruses are present in the U. americanus population or several gammaherpesviruses exist in ursine species.
Keywords: Black bears; DNA sequencing; Degenerative PCR; PCR; UrHV-1; Virus isolation.
Copyright © 2018 Elsevier B.V. All rights reserved.
Figures







References
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Axthelm M.K., Bourdette D.N., Marracci G.H., Su W., Mullaney E.T., Manoharan M., Kohama S.G., Pollaro J., Witkowski E., Wang P., Rooney W.D., Sherman L.S., Wong S.W. Japanese macaque encephalomyelitis: a spontaneous multiple sclerosis-like disease in a nonhuman primate. Annals Neurol. 2011;70:362–373. - PMC - PubMed
-
- Brenner J., Perl S., Lahav D., Garazi S., Oved Z., Shlosberg A., David D. An unusual outbreak of malignant catarrhal fever in a beef herd in Israel. J. Vet. Med. B Infect. Dis. Vet. Public Health. 2002;49:304–307. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

