Online Parallel Accumulation-Serial Fragmentation (PASEF) with a Novel Trapped Ion Mobility Mass Spectrometer
- PMID: 30385480
- PMCID: PMC6283298
- DOI: 10.1074/mcp.TIR118.000900
Online Parallel Accumulation-Serial Fragmentation (PASEF) with a Novel Trapped Ion Mobility Mass Spectrometer
Abstract
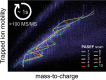
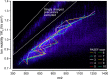
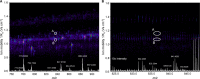
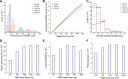
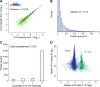
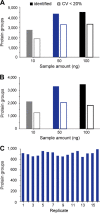
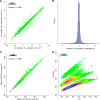
In bottom-up proteomics, peptides are separated by liquid chromatography with elution peak widths in the range of seconds, whereas mass spectra are acquired in about 100 microseconds with time-of-flight (TOF) instruments. This allows adding ion mobility as a third dimension of separation. Among several formats, trapped ion mobility spectrometry (TIMS) is attractive because of its small size, low voltage requirements and high efficiency of ion utilization. We have recently demonstrated a scan mode termed parallel accumulation - serial fragmentation (PASEF), which multiplies the sequencing speed without any loss in sensitivity (Meier et al., PMID: 26538118). Here we introduce the timsTOF Pro instrument, which optimally implements online PASEF. It features an orthogonal ion path into the ion mobility device, limiting the amount of debris entering the instrument and making it very robust in daily operation. We investigate different precursor selection schemes for shotgun proteomics to optimally allocate in excess of 100 fragmentation events per second. More than 600,000 fragmentation spectra in standard 120 min LC runs are achievable, which can be used for near exhaustive precursor selection in complex mixtures or accumulating the signal of weak precursors. In 120 min single runs of HeLa digest, MaxQuant identified more than 6,000 proteins without matching to a library and with high quantitative reproducibility (R > 0.97). Online PASEF achieves a remarkable sensitivity with more than 2,500 proteins identified in 30 min runs of only 10 ng HeLa digest. We also show that highly reproducible collisional cross sections can be acquired on a large scale (R > 0.99). PASEF on the timsTOF Pro is a valuable addition to the technological toolbox in proteomics, with a number of unique operating modes that are only beginning to be explored.
Keywords: Collisional Cross Section; HPLC; Label-free quantification; Mass Spectrometry; PASEF; Protein Identification; Quantification; Sequencing MS; TIMS; Tandem Mass Spectrometry; ion mobility.
© 2018 Meier et al.
Conflict of interest statement
The authors state that they have potential conflicts of interest regarding this work: S.K, H.K. M.L., M.K., N.G., J.D. M.P. and O. R. are employees of Bruker, the manufacturer of the timsTOF Pro. O.H. and N.B. are employees of Evosep. M.M. is an indirect investor in Evosep.
Figures

References
-
- Altelaar A. F. M., Munoz J., and Heck A. J. R. (2012) Next-generation proteomics: towards an integrative view of proteome dynamics. Nat. Rev. Genet. 14, 35–48 - PubMed
-
- Larance M., and Lamond A. I. (2015) Multidimensional proteomics for cell biology. Nat. Rev. Mol. Cell Biol. 16, 269–280 - PubMed
-
- Aebersold R., and Mann M. (2016) Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355 - PubMed
-
- Bekker-Jensen D. B., Kelstrup C. D., Batth T. S., Larsen S. C., Haldrup C., Bramsen J. B., Sørensen K. D., Høyer S., Ørntoft T. F., Andersen C. L., Nielsen M. L., and Olsen J. V. (2017) An Optimized Shotgun Strategy for the Rapid Generation of Comprehensive Human Proteomes. Cell Syst. 4, 587–599.e4 - PMC - PubMed
-
- Michalski A., Cox J., and Mann M. (2011) More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J. Proteome Res. 10, 1785–1793 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

