Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN
- PMID: 30385546
- PMCID: PMC6326779
- DOI: 10.1126/science.aat0572
Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN
Abstract
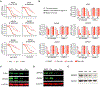
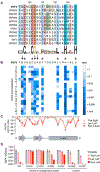
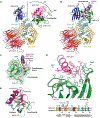
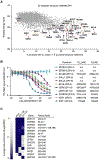
The small molecules thalidomide, lenalidomide, and pomalidomide induce the ubiquitination and proteasomal degradation of the transcription factors Ikaros (IKZF1) and Aiolos (IKZF3) by recruiting a Cys2-His2 (C2H2) zinc finger domain to Cereblon (CRBN), the substrate receptor of the CRL4CRBN E3 ubiquitin ligase. We screened the human C2H2 zinc finger proteome for degradation in the presence of thalidomide analogs, identifying 11 zinc finger degrons. Structural and functional characterization of the C2H2 zinc finger degrons demonstrates how diverse zinc finger domains bind the permissive drug-CRBN interface. Computational zinc finger docking and biochemical analysis predict that more than 150 zinc fingers bind the drug-CRBN complex in vitro, and we show that selective zinc finger degradation can be achieved through compound modifications. Our results provide a rationale for therapeutically targeting transcription factors that were previously considered undruggable.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures


Comment in
-
Evolving Rules for Protein Degradation? Insights from the Zinc Finger Degrome.Biochemistry. 2019 Feb 19;58(7):861-864. doi: 10.1021/acs.biochem.8b01307. Epub 2019 Jan 25. Biochemistry. 2019. PMID: 30681837 No abstract available.
References
-
- Matyskiela ME et al., A novel cereblon modulator recruits GSPT1 to the CRL4CRBN ubiquitin ligase. Nature, 1–24 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

