Recursive Paleohexaploidization Shaped the Durian Genome
- PMID: 30385647
- PMCID: PMC6324235
- DOI: 10.1104/pp.18.00921
Recursive Paleohexaploidization Shaped the Durian Genome
Abstract
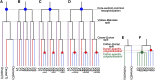
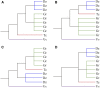
The durian (Durio zibethinus) genome has recently become available, and analysis of this genome reveals two paleopolyploidization events previously inferred as shared with cotton (Gossypium spp.). Here, we reanalyzed the durian genome in comparison with other well-characterized genomes. We found that durian and cotton were actually affected by different polyploidization events: hexaploidization in durian ∼19-21 million years ago (mya) and decaploidization in cotton ∼13-14 mya. Previous interpretations of shared polyploidization events may have resulted from the elevated evolutionary rates in cotton genes due to the decaploidization and insufficient consideration of the complexity of plant genomes. The decaploidization elevated evolutionary rates of cotton genes by ∼64% compared to durian and explained a previous ∼4-fold over dating of the event. In contrast, the hexaploidization in durian did not prominently elevate gene evolutionary rates, likely due to its long generation time. Moreover, divergent evolutionary rates probably explain 98.4% of reconstructed phylogenetic trees of homologous genes being incongruent with expected topology. The findings provide further insight into the roles played by polypoidization in the evolution of genomes and genes, and they suggest revisiting existing reconstructed phylogenetic trees.
© 2019 The author(s). All Rights Reserved.
Figures



Similar articles
-
The draft genome of tropical fruit durian (Durio zibethinus).Nat Genet. 2017 Nov;49(11):1633-1641. doi: 10.1038/ng.3972. Epub 2017 Oct 9. Nat Genet. 2017. PMID: 28991254
-
Upgraded durian genome reveals the role of chromosome reshuffling during ancestral karyotype evolution, lignin biosynthesis regulation, and stress tolerance.Sci China Life Sci. 2024 Jun;67(6):1266-1279. doi: 10.1007/s11427-024-2580-3. Epub 2024 May 17. Sci China Life Sci. 2024. PMID: 38763999
-
Assembly of the durian chloroplast genome using long PacBio reads.Sci Rep. 2020 Oct 7;10(1):15980. doi: 10.1038/s41598-020-73549-4. Sci Rep. 2020. PMID: 33028920 Free PMC article.
-
Sequencing Multiple Cotton Genomes Reveals Complex Structures and Lays Foundation for Breeding.Front Plant Sci. 2020 Sep 16;11:560096. doi: 10.3389/fpls.2020.560096. eCollection 2020. Front Plant Sci. 2020. PMID: 33042184 Free PMC article. Review.
-
Exploring the potential nutraceutical values of durian (Durio zibethinus L.) - an exotic tropical fruit.Food Chem. 2015 Feb 1;168:80-9. doi: 10.1016/j.foodchem.2014.07.020. Epub 2014 Jul 11. Food Chem. 2015. PMID: 25172686 Review.
Cited by
-
PolyReco: A Method to Automatically Label Collinear Regions and Recognize Polyploidy Events Based on the K S Dotplot.Front Genet. 2022 Apr 20;13:842387. doi: 10.3389/fgene.2022.842387. eCollection 2022. Front Genet. 2022. PMID: 35518356 Free PMC article.
-
Resequencing of durian genomes reveals large genetic variations among different cultivars.Front Plant Sci. 2023 Feb 16;14:1137077. doi: 10.3389/fpls.2023.1137077. eCollection 2023. Front Plant Sci. 2023. PMID: 36875624 Free PMC article.
-
Illegitimate Recombination Between Homeologous Genes in Wheat Genome.Front Plant Sci. 2020 Jul 21;11:1076. doi: 10.3389/fpls.2020.01076. eCollection 2020. Front Plant Sci. 2020. PMID: 32849677 Free PMC article.
-
Comparative genomics approach to infer ancestral cell karyotypes and reconstruct the evolutionary trajectories of plant chromosomes.Nat Protoc. 2025 Aug 1. doi: 10.1038/s41596-025-01173-5. Online ahead of print. Nat Protoc. 2025. PMID: 40750714 Review.
-
The genome of giant waterlily provides insights into the origin of angiosperms, leaf gigantism, and stamen function innovation.Plant Commun. 2025 Jun 9;6(6):101342. doi: 10.1016/j.xplc.2025.101342. Epub 2025 Apr 16. Plant Commun. 2025. PMID: 40247621 Free PMC article.
References
-
- Abrouk M, Murat F, Pont C, Messing J, Jackson S, Faraut T, Tannier E, Plomion C, Cooke R, Feuillet C, Salse J (2010) Palaeogenomics of plants: Synteny-based modelling of extinct ancestors. Trends Plant Sci 15: 479–487 - PubMed
-
- Argout X, Salse J, Aury JM, Guiltinan MJ, Droc G, Gouzy J, Allegre M, Chaparro C, Legavre T, Maximova SN, Abrouk M, Murat F, et al. (2011) The genome of Theobroma cacao. Nat Genet 43: 101–108 - PubMed
-
- Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 - PubMed
-
- Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, Corréa M, Da Silva C, et al. (2014) Plant genetics: Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345: 950–953 - PubMed
-
- Charon C, Bruggeman Q, Thareau V, Henry Y (2012) Gene duplication within the Green Lineage: The case of TEL genes. J Exp Bot 63: 5061–5077 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

