Transcriptomic heterogeneity in multifocal prostate cancer
- PMID: 30385730
- PMCID: PMC6238741
- DOI: 10.1172/jci.insight.123468
Transcriptomic heterogeneity in multifocal prostate cancer
Abstract
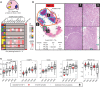
Background: Commercial gene expression assays are guiding clinical decision making in patients with prostate cancer, particularly when considering active surveillance. Given heterogeneity and multifocality of primary prostate cancer, such assays should ideally be robust to the coexistence of unsampled higher grade disease elsewhere in the prostate in order to have clinical utility. Herein, we comprehensively evaluated transcriptomic profiles of primary multifocal prostate cancer to assess robustness to clinically relevant multifocality.
Methods: We designed a comprehensive, multiplexed targeted RNA-sequencing assay capable of assessing multiple transcriptional classes and deriving commercially available prognostic signatures, including the Myriad Prolaris Cell Cycle Progression score, the Oncotype DX Genomic Prostate Score, and the GenomeDX Decipher Genomic Classifier. We applied this assay to a retrospective, multi-institutional cohort of 156 prostate cancer samples. Derived commercial biomarker scores for 120 informative primary prostate cancer samples from 44 cases were determined and compared.
Results: Derived expression scores were positively correlated with tumor grade (rS = 0.53-0.73; all P < 0.001), both within the same case and across the entire cohort. In cases of extreme grade-discordant multifocality (co-occurrence of grade group 1 [GG1] and ≥GG4 foci], gene expression scores were significantly lower in low- (GG1) versus high-grade (≥GG4) foci (all P < 0.001). No significant differences in expression scores, however, were observed between GG1 foci from prostates with and without coexisting higher grade cancer (all P > 0.05).
Conclusions: Multifocal, low-grade and high-grade prostate cancer foci exhibit distinct prognostic expression signatures. These findings demonstrate that prognostic RNA expression assays performed on low-grade prostate cancer biopsy tissue may not provide meaningful information on the presence of coexisting unsampled aggressive disease.
Funding: Prostate Cancer Foundation, National Institutes of Health (U01 CA214170, R01 CA183857, University of Michigan Prostate Specialized Program of Research Excellence [S.P.O.R.E.] P50 CA186786-05, Weill Cornell Medicine S.P.O.R.E. P50 CA211024-01A1), Men of Michigan Prostate Cancer Research Fund, University of Michigan Comprehensive Cancer Center core grant (2-P30-CA-046592-24), A. Alfred Taubman Biomedical Research Institute, and Department of Defense.
Keywords: Cancer; Molecular diagnosis; Oncology; Prostate cancer.
Conflict of interest statement
Figures



Comment in
-
Re: Transcriptomic Heterogeneity in Multifocal Prostate Cancer.J Urol. 2019 Apr;201(4):666-667. doi: 10.1097/JU.0000000000000101. J Urol. 2019. PMID: 30653002 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

