Conservation of the "Outside-in" Germination Pathway in Paraclostridium bifermentans
- PMID: 30386321
- PMCID: PMC6199464
- DOI: 10.3389/fmicb.2018.02487
Conservation of the "Outside-in" Germination Pathway in Paraclostridium bifermentans
Abstract
Clostridium difficile spore germination is initiated in response to certain bile acids and amino acids (e.g., glycine). Though the amino acid-recognizing germinant receptor is unknown, the bile acid germinant receptor is the germination-specific, subtilisin-like pseudoprotease, CspC. In C. difficile the CspB, CspA, and CspC proteins are involved in spore germination. Of these, only CspB is predicted to have catalytic activity because the residues important for catalysis are mutated in the cspA and cspC sequence. The CspB, CspA, and CspC proteins are likely localized to the outer layers of the spore (e.g., the cortex or the coat layers) and not the inner membrane where the Ger-type germinant receptors are located. In C. difficile, germination proceeds in an "outside-in" direction, instead of the "'inside-out" direction observed during the germination of Bacillus subtilis spores. During C. difficile spore germination, cortex fragments are released prior to the release of 2,4-dipicolinic acid (DPA) from the spore core. This is opposite to what occurs during B. subtilis spore germination. To understand if the mechanism C. difficile spore germination is unique or if spores from other organisms germinate in a similar fashion, we analyzed the germination of Paraclostridium bifermentans spores. We find that P. bifermentans spores release cortex fragments prior to DPA during germination and the DPA release from the P. bifermentans spore core can be blocked by high concentrations of osmolytes. Moreover, we find that P. bifermentans spores do not respond to steroid-like compounds (unlike the related C. difficile and P. sordellii organisms), indicating that the mere presence of the Csp proteins does permit germination in response to steroid compounds. Our findings indicate that the "outside in" mechanism of spore germination observed in C. difficile can be found in other bacteria suggesting that this mechanism is a novel pathway for endospore germination.
Keywords: Clostridium; DPA; cortex; germination; spore.
Figures
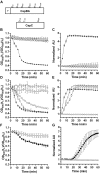
 50 mM
50 mM  50 mM
50 mM  (D,E). Spores alone,
(D,E). Spores alone,  50 mM
50 mM  50 mM
50 mM  50 mM
50 mM  (F,G) Spores alone,
(F,G) Spores alone,  50 mM
50 mM  Data points represent the average from three independent experiments and error bars represent the standard deviation from the mean.
Data points represent the average from three independent experiments and error bars represent the standard deviation from the mean.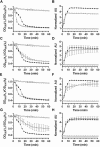
 50 mM
50 mM  50 mM
50 mM 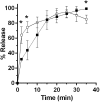
 and for DPA release
and for DPA release  . Data points represent the average from three independent experiments and error bars represent the standard deviation from the mean. Statistical significant was determined using a two-way ANOVA with Sidak’s multiple comparisons test. The asterisk marked points indicate statistical significance (p-value < 0.05).
. Data points represent the average from three independent experiments and error bars represent the standard deviation from the mean. Statistical significant was determined using a two-way ANOVA with Sidak’s multiple comparisons test. The asterisk marked points indicate statistical significance (p-value < 0.05).
 and in absence of
and in absence of  sorbitol. (A,B) 10% sorbitol; (C,D) 20% sorbitol; (E,F) 30% sorbitol; (G,H) 38% sorbitol. Data points represent the average from three independent experiments and error bars represent the standard deviation from the mean.
sorbitol. (A,B) 10% sorbitol; (C,D) 20% sorbitol; (E,F) 30% sorbitol; (G,H) 38% sorbitol. Data points represent the average from three independent experiments and error bars represent the standard deviation from the mean.
Similar articles
-
Revisiting the Role of Csp Family Proteins in Regulating Clostridium difficile Spore Germination.J Bacteriol. 2017 Oct 17;199(22):e00266-17. doi: 10.1128/JB.00266-17. Print 2017 Nov 15. J Bacteriol. 2017. PMID: 28874406 Free PMC article.
-
Dipicolinic Acid Release by Germinating Clostridium difficile Spores Occurs through a Mechanosensing Mechanism.mSphere. 2016 Dec 14;1(6):e00306-16. doi: 10.1128/mSphere.00306-16. eCollection 2016 Nov-Dec. mSphere. 2016. PMID: 27981237 Free PMC article.
-
Spore Cortex Hydrolysis Precedes Dipicolinic Acid Release during Clostridium difficile Spore Germination.J Bacteriol. 2015 Jul;197(14):2276-83. doi: 10.1128/JB.02575-14. Epub 2015 Apr 27. J Bacteriol. 2015. PMID: 25917906 Free PMC article.
-
Terbium chloride influences Clostridium difficile spore germination.Anaerobe. 2019 Aug;58:80-88. doi: 10.1016/j.anaerobe.2019.03.016. Epub 2019 Mar 26. Anaerobe. 2019. PMID: 30926439 Free PMC article. Review.
-
A Revised Understanding of Clostridioides difficile Spore Germination.Trends Microbiol. 2020 Sep;28(9):744-752. doi: 10.1016/j.tim.2020.03.004. Epub 2020 Apr 23. Trends Microbiol. 2020. PMID: 32781028 Review.
Cited by
-
Factors and Conditions That Impact Electroporation of Clostridioides difficile Strains.mSphere. 2020 Mar 4;5(2):e00941-19. doi: 10.1128/mSphere.00941-19. mSphere. 2020. PMID: 32132157 Free PMC article.
-
The impact of YabG mutations on C. difficile spore germination and processing of spore substrates.bioRxiv [Preprint]. 2024 Jun 10:2024.06.10.598338. doi: 10.1101/2024.06.10.598338. bioRxiv. 2024. Update in: Mol Microbiol. 2024 Oct;122(4):534-548. doi: 10.1111/mmi.15316. PMID: 38915615 Free PMC article. Updated. Preprint.
-
Clostridium septicum manifests a bile salt germinant response mediated by Clostridioides difficile csp gene orthologs.Commun Biol. 2024 Aug 6;7(1):947. doi: 10.1038/s42003-024-06617-4. Commun Biol. 2024. PMID: 39103440 Free PMC article.
-
The Impact of YabG Mutations on Clostridioides difficile Spore Germination and Processing of Spore Substrates.Mol Microbiol. 2024 Oct;122(4):534-548. doi: 10.1111/mmi.15316. Epub 2024 Sep 11. Mol Microbiol. 2024. PMID: 39258427
-
Inhibitory effect of licorice extract on the germination and outgrowth of Paraclostridium bifermentans spores.Front Microbiol. 2022 Dec 2;13:1076144. doi: 10.3389/fmicb.2022.1076144. eCollection 2022. Front Microbiol. 2022. PMID: 36532483 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

