The structure of PghL hydrolase bound to its substrate poly-γ-glutamate
- PMID: 30387270
- PMCID: PMC6506827
- DOI: 10.1111/febs.14688
The structure of PghL hydrolase bound to its substrate poly-γ-glutamate
Abstract
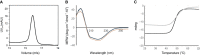
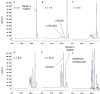
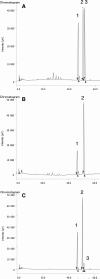
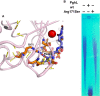
The identification of new strategies to fight bacterial infections in view of the spread of multiple resistance to antibiotics has become mandatory. It has been demonstrated that several bacteria develop poly-γ-glutamic acid (γ-PGA) capsules as a protection from external insults and/or host defence systems. Among the pathogens that shield themselves in these capsules are Bacillus anthracis, Francisella tularensis and several Staphylococcus strains. These are important pathogens with a profound influence on human health. The recently characterised γ-PGA hydrolases, which can dismantle the γ-PGA-capsules, are an attractive new direction that can offer real hope for the development of alternatives to antibiotics, particularly in cases of multidrug resistant bacteria. We have characterised in detail the cleaving mechanism and stereospecificity of the enzyme PghL (previously named YndL) from Bacillus subtilis encoded by a gene of phagic origin and dramatically efficient in degrading the long polymeric chains of γ-PGA. We used X-ray crystallography to solve the three-dimensional structures of the enzyme in its zinc-free, zinc-bound and complexed forms. The protein crystallised with a γ-PGA hexapeptide substrate and thus reveals details of the interaction which could explain the stereospecificity observed and give hints on the catalytic mechanism of this class of hydrolytic enzymes.
Keywords: PGA-hydrolase; antimicrobial drug; biofilm inhibitor; poly-γ-glutamate; virulence.
© 2018 The Authors. The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

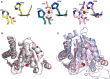
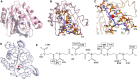
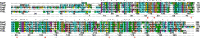
References
-
- Candela T & Fouet A (2006) Poly‐gamma‐glutamate in bacteria. Mol Microbiol 60, 1091–1098. - PubMed
-
- Jang J, Cho M, Lee H, Cha K, Chun J, Hong K, Park J & Rhie G (2013) Monoclonal antibody against the poly‐gamma‐D‐glutamic acid capsule of Bacillus anthracis protects mice from enhanced lethal toxin activity due to capsule and anthrax spore challenge. Biochim Biophys Acta 1830, 2804–2812. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

