US Immigration Westernizes the Human Gut Microbiome
- PMID: 30388453
- PMCID: PMC6498444
- DOI: 10.1016/j.cell.2018.10.029
US Immigration Westernizes the Human Gut Microbiome
Abstract
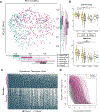
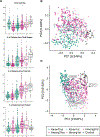
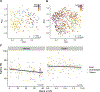
Many US immigrant populations develop metabolic diseases post immigration, but the causes are not well understood. Although the microbiome plays a role in metabolic disease, there have been no studies measuring the effects of US immigration on the gut microbiome. We collected stool, dietary recalls, and anthropometrics from 514 Hmong and Karen individuals living in Thailand and the United States, including first- and second-generation immigrants and 19 Karen individuals sampled before and after immigration, as well as from 36 US-born European American individuals. Using 16S and deep shotgun metagenomic DNA sequencing, we found that migration from a non-Western country to the United States is associated with immediate loss of gut microbiome diversity and function in which US-associated strains and functions displace native strains and functions. These effects increase with duration of US residence and are compounded by obesity and across generations.
Keywords: Bacteriodes; Prevotella; immigrant health; immigration; metagenomics; microbiome; microbiota; obesity; refugee health.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests
D.K. serves as CEO and holds equity in CoreBiome, a company involved in the commercialization of microbiome analysis. The University of Minnesota also has financial interests in CoreBiome under the terms of a license agreement with CoreBiome. These interests have been reviewed and managed by the University of Minnesota in accordance with its Conflict-of-Interest policies.
Figures




Comment in
-
Life in a distant land triggers upheaval in immigrants' microbiomes.Nature. 2018 Nov;563(7730):158. doi: 10.1038/d41586-018-07223-1. Nature. 2018. PMID: 30401844 No abstract available.
-
A Microbiota Assimilation.Cell Metab. 2018 Nov 6;28(5):675-677. doi: 10.1016/j.cmet.2018.10.010. Cell Metab. 2018. PMID: 30403987
References
-
- Al-Ghalith G, and Knights D (2017). BURST enables optimal exhaustive DNA alignment for big data (Zenodo).
-
- Al-Ghalith G, and Knights D (2018). aKronyMer enables database-free metagenome comparison (Zenodo).
-
- Al-Ghalith GA, Vangay P, and Knights D (2015). The guts of obesity: progress and challenges in linking gut microbes to obesity. Discov. Med 19, 81–88. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

