Gesicle-Mediated Delivery of CRISPR/Cas9 Ribonucleoprotein Complex for Inactivating the HIV Provirus
- PMID: 30389355
- PMCID: PMC6318701
- DOI: 10.1016/j.ymthe.2018.10.002
Gesicle-Mediated Delivery of CRISPR/Cas9 Ribonucleoprotein Complex for Inactivating the HIV Provirus
Abstract
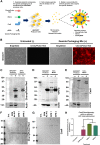
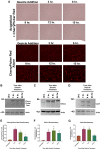
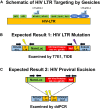
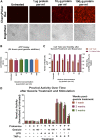
Investigators have utilized the CRISPR/Cas9 gene-editing system to specifically target well-conserved regions of HIV, leading to decreased infectivity and pathogenesis in vitro and ex vivo. We utilized a specialized extracellular vesicle termed a "gesicle" to efficiently, yet transiently, deliver Cas9 in a ribonucleoprotein form targeting the HIV long terminal repeat (LTR). Gesicles are produced through expression of vesicular stomatitis virus glycoprotein and package protein as their cargo, thus bypassing the need for transgene delivery, and allowing finer control of Cas9 expression. Using both NanoSight particle and western blot analysis, we verified production of Cas9-containing gesicles by HEK293FT cells. Application of gesicles to CHME-5 microglia resulted in rapid but transient transfer of Cas9 by western blot, which is present at 1 hr, but is undetectable by 24 hr post-treatment. Gesicle delivery of Cas9 protein preloaded with guide RNA targeting the HIV LTR to HIV-NanoLuc CHME-5 cells generated mutations within the LTR region and copy number loss. Finally, we demonstrated that this treatment resulted in reduced proviral activity under basal conditions and after stimulation with pro-inflammatory factors lipopolysaccharide (LPS) or tumor necrosis factor alpha (TNF-α). These data suggest that gesicles are a viable alternative approach to deliver CRISPR/Cas9 technology.
Keywords: Cas9 ribonucleoprotein; HIV; gesicle.
Copyright © 2018 The American Society of Gene and Cell Therapy. All rights reserved.
Figures



References
-
- Salmaninejad A., Valilou S.F., Bayat H., Ebadi N., Daraei A., Yousefi M., Nesaei A., Mojarrad M. Duchenne muscular dystrophy: an updated review of common available therapies. Int. J. Neurosci. 2018;128:854–864. - PubMed
-
- Schwank G., Koo B.K., Sasselli V., Dekkers J.F., Heo I., Demircan T., Sasaki N., Boymans S., Cuppen E., van der Ent C.K. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

