Expression of murine muscle-enriched A-type lamin-interacting protein (MLIP) is regulated by tissue-specific alternative transcription start sites
- PMID: 30389785
- PMCID: PMC6314141
- DOI: 10.1074/jbc.RA118.003758
Expression of murine muscle-enriched A-type lamin-interacting protein (MLIP) is regulated by tissue-specific alternative transcription start sites
Abstract
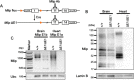
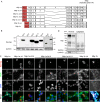
Muscle-enriched lamin-interacting protein (Mlip) is an alternatively spliced gene whose splicing specificity is dictated by tissue type. MLIP is most abundantly expressed in brain, cardiac, and skeletal muscle. In the present study, we systematically mapped the transcriptional start and stop sites of murine Mlip Rapid amplification of cDNA ends (RACE) of Mlip transcripts from the brain, heart, and skeletal muscle revealed two transcriptional start sites (TSSs), exon 1a and exon 1b, and only one transcriptional termination site. RT-PCR analysis of the usage of the two identified TSSs revealed that the heart utilizes only exon 1a for MLIP expression, whereas the brain exclusively uses exon 1b TSS. Loss of Mlip exon 1a in mice resulted in a 7-fold increase in the prevalence of centralized nuclei in muscle fibers with the Mlip exon1a-deficient satellite cells on single fibers exhibiting a significant delay in commitment to a MYOD-positive phenotype. Furthermore, we demonstrate that the A-type lamin-binding domain in MLIP is encoded in exon 1a, indicating that MLIP isoforms generated with exon 1b TSS lack the A-type lamin-binding domain. Collectively these findings suggest that Mlip tissue-specific expression and alternative splicing play a critical role in determining MLIP's functions in mice.
Keywords: MLIP; alternative splicing; brain; cellular localization; gene expression; heart; intracellular trafficking; muscle-enriched lamin-interacting protein; protein isoform; protein-protein interaction; skeletal muscle; splice variant; tissue-specific expression; transcriptional regulation; transcriptional start site.
© 2018 Cattin et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


Similar articles
-
MLIP and Its Potential Influence on Key Oncogenic Pathways.Cells. 2024 Jun 26;13(13):1109. doi: 10.3390/cells13131109. Cells. 2024. PMID: 38994962 Free PMC article. Review.
-
Identification of a novel muscle A-type lamin-interacting protein (MLIP).J Biol Chem. 2011 Jun 3;286(22):19702-13. doi: 10.1074/jbc.M110.165548. Epub 2011 Apr 15. J Biol Chem. 2011. PMID: 21498514 Free PMC article.
-
Muscle Enriched Lamin Interacting Protein (Mlip) Binds Chromatin and Is Required for Myoblast Differentiation.Cells. 2021 Mar 10;10(3):615. doi: 10.3390/cells10030615. Cells. 2021. PMID: 33802236 Free PMC article.
-
Nesprins: tissue-specific expression of epsilon and other short isoforms.PLoS One. 2014 Apr 9;9(4):e94380. doi: 10.1371/journal.pone.0094380. eCollection 2014. PLoS One. 2014. PMID: 24718612 Free PMC article.
-
MLIP-Associated Myopathy: A Case Report and Review of the Literature.J Neuromuscul Dis. 2023;10(2):293-299. doi: 10.3233/JND-221520. J Neuromuscul Dis. 2023. PMID: 36641683 Review.
Cited by
-
Novel homozygous nonsense mutation of MLIP and compensatory alternative splicing.NPJ Genom Med. 2022 Jun 7;7(1):36. doi: 10.1038/s41525-022-00307-y. NPJ Genom Med. 2022. PMID: 35672413 Free PMC article.
-
MLIP causes recessive myopathy with rhabdomyolysis, myalgia and baseline elevated serum creatine kinase.Brain. 2021 Oct 22;144(9):2722-2731. doi: 10.1093/brain/awab275. Brain. 2021. PMID: 34581780 Free PMC article.
-
MLIP genotype as a predictor of pharmacological response in primary open-angle glaucoma and ocular hypertension.Sci Rep. 2021 Jan 15;11(1):1583. doi: 10.1038/s41598-020-80954-2. Sci Rep. 2021. PMID: 33452295 Free PMC article. Clinical Trial.
-
MLIP and Its Potential Influence on Key Oncogenic Pathways.Cells. 2024 Jun 26;13(13):1109. doi: 10.3390/cells13131109. Cells. 2024. PMID: 38994962 Free PMC article. Review.
-
MicroRNA205: A Key Regulator of Cardiomyocyte Transition from Proliferative to Hypertrophic Growth in the Neonatal Heart.Int J Mol Sci. 2024 Feb 12;25(4):2206. doi: 10.3390/ijms25042206. Int J Mol Sci. 2024. PMID: 38396885 Free PMC article.
References
-
- Cattin M.-E., Wang J., Weldrick J. J., Roeske C. L., Mak E., Thorn S. L., DaSilva J. N., Wang Y., Lusis A. J., and Burgon P. G. (2015) Deletion of MLIP (muscle-enriched A-type lamin-interacting protein) leads to cardiac hyperactivation of Akt/mammalian target of rapamycin (mTOR) and impaired cardiac adaptation. J. Biol. Chem. 290, 26699–26714 10.1074/jbc.M115.678433 - DOI - PMC - PubMed
-
- Huang Z.-P., Kataoka M., Chen J., Wu G., Ding J., Nie M., Lin Z., Liu J., Hu X., Ma L., Zhou B., Wakimoto H., Zeng C., Kyselovic J., Deng Z.-L., et al. (2015) Cardiomyocyte-enriched protein CIP protects against pathophysiological stresses and regulates cardiac homeostasis. J. Clin. Investig. 125, 4122–4134 10.1172/JCI82423 - DOI - PMC - PubMed
-
- Esslinger U., Garnier S., Korniat A., Proust C., Kararigas G., Müller-Nurasyid M., Empana J.-P., Morley M. P., Perret C., Stark K., Bick A. G., Prasad S. K., Kriebel J., Li J., Tiret L., et al. (2017) Exome-wide association study reveals novel susceptibility genes to sporadic dilated cardiomyopathy. PLoS One 12, e0172995 10.1371/journal.pone.0172995 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

