Transgenerational Epigenetic Inheritance Is Negatively Regulated by the HERI-1 Chromodomain Protein
- PMID: 30389807
- PMCID: PMC6283161
- DOI: 10.1534/genetics.118.301456
Transgenerational Epigenetic Inheritance Is Negatively Regulated by the HERI-1 Chromodomain Protein
Abstract
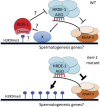
Transgenerational epigenetic inheritance (TEI) is the inheritance of epigenetic information for two or more generations. In most cases, TEI is limited to a small number of generations (two to three). The short-term nature of TEI could be set by innate biochemical limitations to TEI or by genetically encoded systems that actively limit TEI. In Caenorhabditis elegans, double-stranded RNA (dsRNA)-mediated gene silencing [RNAi (RNA interference)] can be inherited (termed RNAi inheritance or RNA-directed TEI). To identify systems that might actively limit RNA-directed TEI, we conducted a forward genetic screen for factors whose mutation enhanced RNAi inheritance. This screen identified the gene heritable enhancer of RNAi (heri-1), whose mutation causes RNAi inheritance to last longer (> 20 generations) than normal. heri-1 encodes a protein with a chromodomain, and a kinase homology domain that is expressed in germ cells and localizes to nuclei. In C. elegans, a nuclear branch of the RNAi pathway [termed the nuclear RNAi or NRDE (nuclear RNA defective) pathway] promotes RNAi inheritance. We find that heri-1(-) animals have defects in spermatogenesis that are suppressible by mutations in the nuclear RNAi Argonaute (Ago) HRDE-1, suggesting that HERI-1 might normally act in sperm progenitor cells to limit nuclear RNAi and/or RNAi inheritance. Consistent with this idea, we find that the NRDE nuclear RNAi pathway is hyperresponsive to experimental RNAi treatments in heri-1 mutant animals. Interestingly, HERI-1 binds to genes targeted by RNAi, suggesting that HERI-1 may have a direct role in limiting nuclear RNAi and, therefore, RNAi inheritance. Finally, the recruitment of HERI-1 to chromatin depends upon the same factors that drive cotranscriptional gene silencing, suggesting that the generational perdurance of RNAi inheritance in C. elegans may be set by competing pro- and antisilencing outputs of the nuclear RNAi machinery.
Keywords: RNAi; chromatin; small RNAs; transgenerational epigenetic inheritance.
Copyright © 2018 by the Genetics Society of America.
Figures



Similar articles
-
A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality.Nature. 2012 Sep 20;489(7416):447-51. doi: 10.1038/nature11352. Epub 2012 Jul 18. Nature. 2012. PMID: 22810588 Free PMC article.
-
A Family of Argonaute-Interacting Proteins Gates Nuclear RNAi.Mol Cell. 2020 Jun 4;78(5):862-875.e8. doi: 10.1016/j.molcel.2020.04.007. Epub 2020 Apr 28. Mol Cell. 2020. PMID: 32348780 Free PMC article.
-
The RNAi Inheritance Machinery of Caenorhabditis elegans.Genetics. 2017 Jul;206(3):1403-1416. doi: 10.1534/genetics.116.198812. Epub 2017 May 22. Genetics. 2017. PMID: 28533440 Free PMC article.
-
Distinct nuclear and cytoplasmic machineries cooperatively promote the inheritance of RNAi in Caenorhabditis elegans.Biol Cell. 2018 Oct;110(10):217-224. doi: 10.1111/boc.201800031. Epub 2018 Sep 9. Biol Cell. 2018. PMID: 30132958 Review.
-
Mechanisms of epigenetic regulation by C. elegans nuclear RNA interference pathways.Semin Cell Dev Biol. 2022 Jul;127:142-154. doi: 10.1016/j.semcdb.2021.11.018. Epub 2021 Dec 6. Semin Cell Dev Biol. 2022. PMID: 34876343 Review.
Cited by
-
Nongenetic inheritance and multigenerational plasticity in the nematode C. elegans.Elife. 2020 Aug 25;9:e58498. doi: 10.7554/eLife.58498. Elife. 2020. PMID: 32840479 Free PMC article. Review.
-
Small RNAs Worm Up Transgenerational Epigenetics Research.DNA (Basel). 2021 Dec;1(2):37-48. doi: 10.3390/dna1020005. Epub 2021 Sep 29. DNA (Basel). 2021. PMID: 34725653 Free PMC article.
-
Heritable epigenetic changes are constrained by the dynamics of regulatory architectures.Elife. 2024 May 8;12:RP92093. doi: 10.7554/eLife.92093. Elife. 2024. PMID: 38717010 Free PMC article.
-
Epigenetic inheritance of gene silencing is maintained by a self-tuning mechanism based on resource competition.Cell Syst. 2023 Jan 18;14(1):24-40.e11. doi: 10.1016/j.cels.2022.12.003. Cell Syst. 2023. PMID: 36657390 Free PMC article.
-
The third barrier to transgenerational inheritance in animals: somatic epigenetic resetting.EMBO Rep. 2023 Apr 5;24(4):e56615. doi: 10.15252/embr.202256615. Epub 2023 Mar 2. EMBO Rep. 2023. PMID: 36862326 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

