Tissue-resident memory T cells populate the human brain
- PMID: 30389931
- PMCID: PMC6214977
- DOI: 10.1038/s41467-018-07053-9
Tissue-resident memory T cells populate the human brain
Abstract
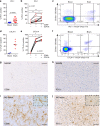
Most tissues are populated by tissue-resident memory T cells (TRM cells), which are adapted to their niche and appear to be indispensable for local protection against pathogens. Here we show that human white matter-derived brain CD8+ T cells can be subsetted into CD103-CD69+ and CD103+CD69+ T cells both with a phenotypic and transcription factor profile consistent with TRM cells. Specifically, CD103 expression in brain CD8+ T cells correlates with reduced expression of differentiation markers, increased expression of tissue-homing chemokine receptors, intermediate and low expression of the transcription factors T-bet and eomes, increased expression of PD-1 and CTLA-4, and low expression of cytolytic enzymes with preserved polyfunctionality upon activation. Brain CD4+ T cells also display TRM cell-associated markers but have low CD103 expression. We conclude that the human brain is surveilled by TRM cells, providing protection against neurotropic virus reactivation, whilst being under tight control of key immune checkpoint molecules.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

