Selective detections of single-viruses using solid-state nanopores
- PMID: 30390013
- PMCID: PMC6214978
- DOI: 10.1038/s41598-018-34665-4
Selective detections of single-viruses using solid-state nanopores
Abstract
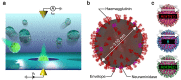
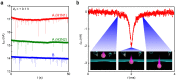
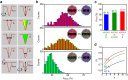
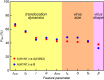
Rapid diagnosis of flu before symptom onsets can revolutionize our health through diminishing a risk for serious complication as well as preventing infectious disease outbreak. Sensor sensitivity and selectivity are key to accomplish this goal as the number of virus is quite small at the early stage of infection. Here we report on label-free electrical diagnostics of influenza based on nanopore analytics that distinguishes individual virions by their distinct physical features. We accomplish selective resistive-pulse sensing of single flu virus having negative surface charges in a physiological media by exploiting electroosmotic flow to filter contaminants at the Si3N4 pore orifice. We demonstrate identifications of allotypes with 68% accuracy at the single-virus level via pattern classifications of the ionic current signatures. We also show that this discriminability becomes >95% under a binomial distribution theorem by ensembling the pulse data of >20 virions. This simple mechanism is versatile for point-of-care tests of a wide range of flu types.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Identifying Single Viruses Using Biorecognition Solid-State Nanopores.J Am Chem Soc. 2018 Dec 5;140(48):16834-16841. doi: 10.1021/jacs.8b10854. Epub 2018 Nov 26. J Am Chem Soc. 2018. PMID: 30475615
-
Identifying Single Particles in Air Using a 3D-Integrated Solid-State Pore.ACS Sens. 2019 Mar 22;4(3):748-755. doi: 10.1021/acssensors.9b00113. Epub 2019 Mar 5. ACS Sens. 2019. PMID: 30788967
-
Discriminating single-bacterial shape using low-aspect-ratio pores.Sci Rep. 2017 Dec 12;7(1):17371. doi: 10.1038/s41598-017-17443-6. Sci Rep. 2017. PMID: 29234023 Free PMC article.
-
Asymmetric ion transport through ion-channel-mimetic solid-state nanopores.Acc Chem Res. 2013 Dec 17;46(12):2834-46. doi: 10.1021/ar400024p. Epub 2013 May 28. Acc Chem Res. 2013. PMID: 23713693 Review.
-
Challenges of Single-Molecule DNA Sequencing with Solid-State Nanopores.Adv Exp Med Biol. 2019;1129:131-142. doi: 10.1007/978-981-13-6037-4_9. Adv Exp Med Biol. 2019. PMID: 30968365 Review.
Cited by
-
Synchronized resistive-pulse analysis with flow visualization for single micro- and nanoscale objects driven by optical vortex in double orifice.Sci Rep. 2021 Apr 29;11(1):9323. doi: 10.1038/s41598-021-87822-7. Sci Rep. 2021. PMID: 33927219 Free PMC article.
-
Nanopore Impedance Spectroscopy Reveals Electrical Properties of Single Nanoparticles for Detecting and Identifying Pathogenic Viruses.ACS Omega. 2023 Apr 6;8(16):14684-14693. doi: 10.1021/acsomega.3c00628. eCollection 2023 Apr 25. ACS Omega. 2023. PMID: 37125101 Free PMC article.
-
Polarization Induced Electro-Functionalization of Pore Walls: A Contactless Technology.Biosensors (Basel). 2019 Oct 11;9(4):121. doi: 10.3390/bios9040121. Biosensors (Basel). 2019. PMID: 31614545 Free PMC article. Review.
-
Focus on using nanopore technology for societal health, environmental, and energy challenges.Nano Res. 2022;15(11):9906-9920. doi: 10.1007/s12274-022-4379-2. Epub 2022 May 20. Nano Res. 2022. PMID: 35610982 Free PMC article. Review.
-
Combination of Single-Molecule Electrical Measurements and Machine Learning for the Identification of Single Biomolecules.ACS Omega. 2020 Jan 7;5(2):959-964. doi: 10.1021/acsomega.9b03660. eCollection 2020 Jan 21. ACS Omega. 2020. PMID: 31984250 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

