MDM2 and mitochondrial function: One complex intersection
- PMID: 30391206
- PMCID: PMC6448155
- DOI: 10.1016/j.bcp.2018.10.032
MDM2 and mitochondrial function: One complex intersection
Abstract
Decades of research reveal that MDM2 participates in cellular processes ranging from macro-molecular metabolism to cancer signaling mechanisms. Two recent studies uncovered a new role for MDM2 in mitochondrial bioenergetics. Through the negative regulation of NDUFS1 (NADH:ubiquinone oxidoreductase 75 kDa Fe-S protein 1) and MT-ND6 (NADH dehydrogenase 6), MDM2 decreases the function and efficiency of Complex I (CI). These observations propose several important questions: (1) Where does MDM2 affect CI activity? (2) What are the cellular consequences of MDM2-mediated regulation of CI? (3) What are the physiological implications of these interactions? Here, we will address these questions and position these observations within the MDM2 literature.
Keywords: Apoptosis; Bioenergetics; Complex I; Electron transport chain; MDM2; Mitochondria; Oncogene; Tumor suppressor; p53.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures
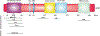
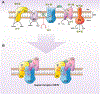

Similar articles
-
MDM2 Integrates Cellular Respiration and Apoptotic Signaling through NDUFS1 and the Mitochondrial Network.Mol Cell. 2019 May 2;74(3):452-465.e7. doi: 10.1016/j.molcel.2019.02.012. Epub 2019 Mar 14. Mol Cell. 2019. PMID: 30879903 Free PMC article.
-
Mitochondrial MDM2 Regulates Respiratory Complex I Activity Independently of p53.Mol Cell. 2018 Feb 15;69(4):594-609.e8. doi: 10.1016/j.molcel.2018.01.023. Mol Cell. 2018. PMID: 29452639 Free PMC article.
-
[MDM2 promotes cell death by interacting with mitochondrial bioenergetics].Med Sci (Paris). 2021 Apr;37(4):397-399. doi: 10.1051/medsci/2021040. Epub 2021 Apr 28. Med Sci (Paris). 2021. PMID: 33908859 French.
-
Mdm2-mediated ubiquitylation: p53 and beyond.Cell Death Differ. 2010 Jan;17(1):93-102. doi: 10.1038/cdd.2009.68. Cell Death Differ. 2010. PMID: 19498444 Review.
-
MDM2 oligomers: antagonizers of the guardian of the genome.Oncogene. 2016 Dec 1;35(48):6157-6165. doi: 10.1038/onc.2016.88. Epub 2016 Apr 4. Oncogene. 2016. PMID: 27041565 Free PMC article. Review.
Cited by
-
MDM2-Mediated Ubiquitination of RXRβ Contributes to Mitochondrial Damage and Related Inflammation in Atherosclerosis.Int J Mol Sci. 2022 May 21;23(10):5766. doi: 10.3390/ijms23105766. Int J Mol Sci. 2022. PMID: 35628577 Free PMC article.
-
Dual inhibitor of MDM2 and NFAT1 for experimental therapy of breast cancer: in vitro and in vivo anticancer activities and newly discovered effects on cancer metabolic pathways.Front Pharmacol. 2025 Feb 19;16:1531667. doi: 10.3389/fphar.2025.1531667. eCollection 2025. Front Pharmacol. 2025. PMID: 40046748 Free PMC article.
-
Impact of supraphysiologic MDM2 expression on chromatin networks and therapeutic responses in sarcoma.Cell Genom. 2023 May 11;3(7):100321. doi: 10.1016/j.xgen.2023.100321. eCollection 2023 Jul 12. Cell Genom. 2023. PMID: 37492096 Free PMC article.
-
Targeting the Metabolic Paradigms in Cancer and Diabetes.Biomedicines. 2024 Jan 17;12(1):211. doi: 10.3390/biomedicines12010211. Biomedicines. 2024. PMID: 38255314 Free PMC article. Review.
-
LncRNA NR_045147 modulates osteogenic differentiation and migration in PDLSCs via ITGB3BP degradation and mitochondrial dysfunction.Stem Cells Transl Med. 2025 Feb 11;14(2):szae088. doi: 10.1093/stcltm/szae088. Stem Cells Transl Med. 2025. PMID: 39674578 Free PMC article.
References
-
- Ahn BY, Trinh DL, Zajchowski LD, Lee B, Elwi AN, and Kim SW (2010). Tid1 is a new regulator of p53 mitochondrial translocation and apoptosis in cancer. Oncogene 29, 1155–1166. - PubMed
-
- Alkhalaf M, Ganguli G, Messaddeq N, Le Meur M, and Wasylyk B (1999). MDM2 overexpression generates a skin phenotype in both wild type and p53 null mice. Oncogene 18, 1419–1434. - PubMed
-
- Benard G, Bellance N, James D, Parrone P, Fernandez H, Letellier T, and Rossignol R (2007). Mitochondrial bioenergetics and structural network organization. J Cell Sci 120, 838–848. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

