Molecular basis for the emergence of a new hospital endemic tigecycline-resistant Enterococcus faecalis ST103 lineage
- PMID: 30393188
- PMCID: PMC6599719
- DOI: 10.1016/j.meegid.2018.10.018
Molecular basis for the emergence of a new hospital endemic tigecycline-resistant Enterococcus faecalis ST103 lineage
Abstract
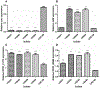
Enterococcus faecalis are a major cause of nosocomial infection worldwide, and the spread of vancomycin resistant strains (VRE) limits treatment options. Tigecycline-resistant VRE began to be isolated from inpatients at a Brazilian hospital within months following the addition of tigecycline to the hospital formulary. This was found to be the result of a spread of an ST103 E. faecalis clone. Our objective was to identify the basis for tigecycline resistance in this lineage. The genomes of two closely related tigecycline-susceptible (MIC = 0.06 mg/L), and three representative tigecycline-resistant (MIC = 1 mg/L) ST103 isolates were sequenced and compared. Further, efforts were undertaken to recapitulate the emergence of resistant strains in vitro. The specific mutations identified in clinical isolates in several cases were within the same genes identified in laboratory-evolved strains. The contribution of various polymorphisms to the resistance phenotype was assessed by trans-complementation of the wild type or mutant alleles, by testing for differences in mRNA abundance, and/or by examining the phenotype of transposon insertion mutants. Among tigecycline-resistant clinical isolates, five genes contained non-synonymous mutations, including two genes known to be related to enterococcal tigecycline resistance (tetM and rpsJ). Finally, within the in vitro-selected resistant variants, mutation in the gene for a MarR-family response regulator was associated with tigecycline resistance. This study shows that E. faecalis mutates to attain tigecycline resistance through the complex interplay of multiple mechanisms, along multiple evolutionary trajectories.
Keywords: Tigecycline resistance; Vancomycin-resistant enterococci; rpsJ; tetM.
Copyright © 2018 Elsevier B.V. All rights reserved.
Conflict of interest statement
Transparency declarations
We declare no conflicts of interest.
Figures

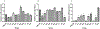
Similar articles
-
Different VanA Elements in E. faecalis and in E. faecium Suggest at Least Two Origins of Tn1546 Among VRE in a Brazilian Hospital.Microb Drug Resist. 2015 Jun;21(3):320-8. doi: 10.1089/mdr.2014.0077. Epub 2015 Jan 14. Microb Drug Resist. 2015. PMID: 25588068
-
Nosocomial infection caused by vancomycin-susceptible multidrug-resistant Enterococcus faecalis over a long period in a university hospital in Japan.Microbiol Immunol. 2014 Nov;58(11):607-14. doi: 10.1111/1348-0421.12190. Microbiol Immunol. 2014. PMID: 25145983
-
Molecular characterization of vancomycin-resistant enterococci isolated from a hospital in Beijing, China.J Microbiol Immunol Infect. 2019 Jun;52(3):433-442. doi: 10.1016/j.jmii.2018.12.008. Epub 2019 Feb 11. J Microbiol Immunol Infect. 2019. PMID: 30827858
-
The emerging problem of linezolid-resistant enterococci.J Glob Antimicrob Resist. 2018 Jun;13:11-19. doi: 10.1016/j.jgar.2017.10.018. Epub 2017 Oct 31. J Glob Antimicrob Resist. 2018. PMID: 29101082
-
Treatment options for vancomycin-resistant enterococcal infections.Drugs. 2002;62(3):425-41. doi: 10.2165/00003495-200262030-00002. Drugs. 2002. PMID: 11827558 Review.
Cited by
-
Profiling Antibiotic Susceptibility among Distinct Enterococcus faecalis Isolates from Dental Root Canals.Antibiotics (Basel). 2023 Dec 24;13(1):18. doi: 10.3390/antibiotics13010018. Antibiotics (Basel). 2023. PMID: 38247577 Free PMC article.
-
Antimicrobial Activity of an Fmoc-Plantaricin 149 Derivative Peptide against Multidrug-Resistant Bacteria.Antibiotics (Basel). 2023 Feb 15;12(2):391. doi: 10.3390/antibiotics12020391. Antibiotics (Basel). 2023. PMID: 36830301 Free PMC article.
-
Exploring Daptomycin Hypersensitivity in Enterococcus faecium: The Impact of LafB Mutation on Bacterial Virulence.Int J Mol Sci. 2025 Jun 20;26(13):5935. doi: 10.3390/ijms26135935. Int J Mol Sci. 2025. PMID: 40649721 Free PMC article.
-
Targeted IS-element sequencing uncovers transposition dynamics during selective pressure in enterococci.PLoS Pathog. 2023 Jun 2;19(6):e1011424. doi: 10.1371/journal.ppat.1011424. eCollection 2023 Jun. PLoS Pathog. 2023. PMID: 37267422 Free PMC article.
-
Mechanism of Eravacycline Resistance in Clinical Enterococcus faecalis Isolates From China.Front Microbiol. 2020 May 25;11:916. doi: 10.3389/fmicb.2020.00916. eCollection 2020. Front Microbiol. 2020. PMID: 32523563 Free PMC article.
References
-
- Agência Nacional de Vigilância Sanitária Registro ANVISA n° 1211002630013 - TYGACIL- VÁLIDO. https://www.smerp.com.br/anvisa/?ac=prodDetail&anvisaId=1211002630013>.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

