Biallelic mutations in DNA ligase 1 underlie a spectrum of immune deficiencies
- PMID: 30395541
- PMCID: PMC6264644
- DOI: 10.1172/JCI99629
Biallelic mutations in DNA ligase 1 underlie a spectrum of immune deficiencies
Abstract
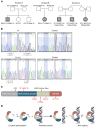
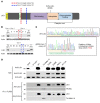
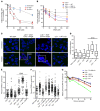
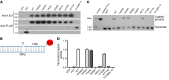
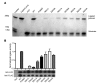
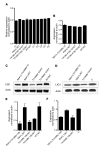
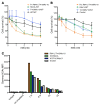
We report the molecular, cellular, and clinical features of 5 patients from 3 kindreds with biallelic mutations in the autosomal LIG1 gene encoding DNA ligase 1. The patients exhibited hypogammaglobulinemia, lymphopenia, increased proportions of circulating γδT cells, and erythrocyte macrocytosis. Clinical severity ranged from a mild antibody deficiency to a combined immunodeficiency requiring hematopoietic stem cell transplantation. Using engineered LIG1-deficient cell lines, we demonstrated chemical and radiation defects associated with the mutant alleles, which variably impaired the DNA repair pathway. We further showed that these LIG1 mutant alleles are amorphic or hypomorphic, and exhibited variably decreased enzymatic activities, which lead to premature release of unligated adenylated DNA. The variability of the LIG1 genotypes in the patients was consistent with that of their immunological and clinical phenotypes. These data suggest that different forms of autosomal recessive, partial DNA ligase 1 deficiency underlie an immunodeficiency of variable severity.
Keywords: B cells; Genetic diseases; Genetics; Immunology; T cell development.
Conflict of interest statement
Figures



Similar articles
-
Severe Combined Immunodeficiency from a Homozygous DNA Ligase 1 Mutant with Reduced Catalytic Activity but Increased Ligation Fidelity.J Clin Immunol. 2024 Jun 19;44(7):151. doi: 10.1007/s10875-024-01754-1. J Clin Immunol. 2024. PMID: 38896336 Free PMC article.
-
Altered DNA ligase activity in human disease.Mutagenesis. 2020 Feb 13;35(1):51-60. doi: 10.1093/mutage/gez026. Mutagenesis. 2020. PMID: 31630206 Free PMC article. Review.
-
Rare variants of DNA ligase 1 show distinct mechanisms of deficiency.J Biol Chem. 2024 Dec;300(12):107957. doi: 10.1016/j.jbc.2024.107957. Epub 2024 Nov 5. J Biol Chem. 2024. PMID: 39510190 Free PMC article.
-
LIG1 syndrome mutations remodel a cooperative network of ligand binding interactions to compromise ligation efficiency.Nucleic Acids Res. 2021 Feb 22;49(3):1619-1630. doi: 10.1093/nar/gkaa1297. Nucleic Acids Res. 2021. PMID: 33444456 Free PMC article.
-
DNA ligase IV syndrome; a review.Orphanet J Rare Dis. 2016 Oct 7;11(1):137. doi: 10.1186/s13023-016-0520-1. Orphanet J Rare Dis. 2016. PMID: 27717373 Free PMC article. Review.
Cited by
-
Severe Combined Immunodeficiency from a Homozygous DNA Ligase 1 Mutant with Reduced Catalytic Activity but Increased Ligation Fidelity.J Clin Immunol. 2024 Jun 19;44(7):151. doi: 10.1007/s10875-024-01754-1. J Clin Immunol. 2024. PMID: 38896336 Free PMC article.
-
Role of circular RNAs in DNA repair.RNA Biol. 2024 Jan;21(1):149-161. doi: 10.1080/15476286.2024.2429945. Epub 2024 Nov 17. RNA Biol. 2024. PMID: 39550713 Free PMC article. Review.
-
A Novel Non-Coding Variant in DCLRE1C Results in Deregulated Splicing and Induces SCID Through the Generation of a Truncated ARTEMIS Protein That Fails to Support V(D)J Recombination and DNA Damage Repair.Front Immunol. 2021 Jun 17;12:674226. doi: 10.3389/fimmu.2021.674226. eCollection 2021. Front Immunol. 2021. PMID: 34220820 Free PMC article.
-
LIG1 Is a Synthetic Lethal Target in BRCA1 Mutant Cancers.Mol Cancer Ther. 2025 Apr 2;24(4):618-627. doi: 10.1158/1535-7163.MCT-24-0598. Mol Cancer Ther. 2025. PMID: 39868490 Free PMC article.
-
Altered DNA ligase activity in human disease.Mutagenesis. 2020 Feb 13;35(1):51-60. doi: 10.1093/mutage/gez026. Mutagenesis. 2020. PMID: 31630206 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

