Pyronaridine exerts potent cytotoxicity on human breast and hematological cancer cells through induction of apoptosis
- PMID: 30395606
- PMCID: PMC6218039
- DOI: 10.1371/journal.pone.0206467
Pyronaridine exerts potent cytotoxicity on human breast and hematological cancer cells through induction of apoptosis
Abstract
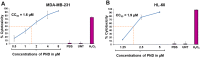
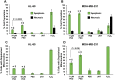
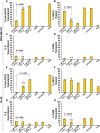
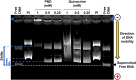
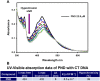
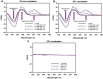
The potent antimalarial drug pyronaridine (PND) was tested for its potential as an anticancer drug. After exposing cancerous (17) and non-cancerous (2) cells to PND for 72 hr, PND was found to exhibit consistent and potent cytotoxic activity at low micromolar (μM) concentrations that ranged from 1.6 μM to 9.4 μM. Moreover, PND exerted a significant selective cytotoxicity index (SCI) on five out of seven breast cancer cell lines tested, with favorable values of 2.5 to 4.4, as compared with the non-cancerous breast MCF-10A cell line. By using the same comparison, PND exhibited a significant SCI on three out of four leukemia/lymphoma cell lines with promising values of 3.3 to 3.5. One breast cancer and one leukemia cell line were tested further in order to determine the likely mode of action of PND. PND was found to consistently elicit phosphatidylserine externalization, mitochondrial depolarization, and DNA fragmentation, in both the triple negative MDA-MB-231 breast cancer and HL-60 leukemia cell lines. In addition, PND treatment altered cell cycle progression in both cancer cells. Subsequent DNA mobility-shift assays, UV-Visible spectroscopic titrations, and circular dichroism (CD) experiments revealed that PND intercalates with DNA. The findings presented in this study indicates that PND induces apoptosis and interfered with cell cycle progression of cancer cell lines and these results indicate that this drug has the potential as a repurposed drug for cancer therapy.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Qi J, Yang CZ, Wang CY, Wang SB, Yang M, Wang JH. Function and mechanism of pyronaridine: a new inhibitor of P-glycoprotein-mediated multidrug resistance. Acta Pharmacol Sin. 2002; 23(6):544–550. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

