Common ancestry of heterodimerizing TALE homeobox transcription factors across Metazoa and Archaeplastida
- PMID: 30396330
- PMCID: PMC6219170
- DOI: 10.1186/s12915-018-0605-5
Common ancestry of heterodimerizing TALE homeobox transcription factors across Metazoa and Archaeplastida
Erratum in
-
Correction to: Common ancestry of heterodimerizing TALE homeobox transcription factors across Metazoa and Archaeplastida.BMC Biol. 2020 Jan 9;18(1):4. doi: 10.1186/s12915-020-0737-2. BMC Biol. 2020. PMID: 31918709 Free PMC article.
Abstract
Background: Complex multicellularity requires elaborate developmental mechanisms, often based on the versatility of heterodimeric transcription factor (TF) interactions. Homeobox TFs in the TALE superclass are deeply embedded in the gene regulatory networks that orchestrate embryogenesis. Knotted-like homeobox (KNOX) TFs, homologous to animal MEIS, have been found to drive the haploid-to-diploid transition in both unicellular green algae and land plants via heterodimerization with other TALE superclass TFs, demonstrating remarkable functional conservation of a developmental TF across lineages that diverged one billion years ago. Here, we sought to delineate whether TALE-TALE heterodimerization is ancestral to eukaryotes.
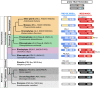
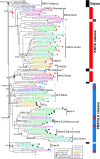
Results: We analyzed TALE endowment in the algal radiations of Archaeplastida, ancestral to land plants. Homeodomain phylogeny and bioinformatics analysis partitioned TALEs into two broad groups, KNOX and non-KNOX. Each group shares previously defined heterodimerization domains, plant KNOX-homology in the KNOX group and animal PBC-homology in the non-KNOX group, indicating their deep ancestry. Protein-protein interaction experiments showed that the TALEs in the two groups all participated in heterodimerization.
Conclusions: Our study indicates that the TF dyads consisting of KNOX/MEIS and PBC-containing TALEs must have evolved early in eukaryotic evolution. Based on our results, we hypothesize that in early eukaryotes, the TALE heterodimeric configuration provided transcription-on switches via dimerization-dependent subcellular localization, ensuring execution of the haploid-to-diploid transition only when the gamete fusion is correctly executed between appropriate partner gametes. The TALE switch then diversified in the several lineages that engage in a complex multicellular organization.
Keywords: Archaeplastida evolution; Developmental mechanism; KNOX transcription factor; PBC-homology; TALE-class homeobox; Transcription factor heterodimerization.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals.Nucleic Acids Res. 1997 Nov 1;25(21):4173-80. doi: 10.1093/nar/25.21.4173. Nucleic Acids Res. 1997. PMID: 9336443 Free PMC article.
-
The PBC domain contains a MEINOX domain: coevolution of Hox and TALE homeobox genes?Dev Genes Evol. 1998 Apr;208(2):113-6. doi: 10.1007/s004270050161. Dev Genes Evol. 1998. PMID: 9569353
-
Ancestry and diversity of BEL1-like homeobox genes revealed by gymnosperm ( Gnetum gnemon) homologs.Dev Genes Evol. 2002 Oct;212(9):452-7. doi: 10.1007/s00427-002-0259-7. Epub 2002 Jul 31. Dev Genes Evol. 2002. PMID: 12373591
-
KNOX homeobox genes potentially have similar function in both diploid unicellular and multicellular meristems, but not in haploid meristems.Evol Dev. 2005 Jan-Feb;7(1):69-78. doi: 10.1111/j.1525-142X.2005.05008.x. Evol Dev. 2005. PMID: 15642091 Review.
-
Knots in the family tree: evolutionary relationships and functions of knox homeobox genes.Plant Mol Biol. 2000 Jan;42(1):151-66. Plant Mol Biol. 2000. PMID: 10688134 Review.
Cited by
-
The transcriptional regulator MEIS2 sets up the ground state for palatal osteogenesis in mice.J Biol Chem. 2020 Apr 17;295(16):5449-5460. doi: 10.1074/jbc.RA120.012684. Epub 2020 Mar 13. J Biol Chem. 2020. PMID: 32169905 Free PMC article.
-
Gamete expression of TALE class HD genes activates the diploid sporophyte program in Marchantia polymorpha.Elife. 2021 Sep 17;10:e57088. doi: 10.7554/eLife.57088. Elife. 2021. PMID: 34533136 Free PMC article.
-
Life cycle and functional genomics of the unicellular red alga Galdieria for elucidating algal and plant evolution and industrial use.Proc Natl Acad Sci U S A. 2022 Oct 11;119(41):e2210665119. doi: 10.1073/pnas.2210665119. Epub 2022 Oct 4. Proc Natl Acad Sci U S A. 2022. PMID: 36194630 Free PMC article.
-
Enhanced sensitivity of TAPscan v4 enables comprehensive analysis of streptophyte transcription factor evolution.Plant J. 2025 Jan;121(1):e17184. doi: 10.1111/tpj.17184. Epub 2024 Dec 12. Plant J. 2025. PMID: 39666589 Free PMC article.
-
Correction to: Common ancestry of heterodimerizing TALE homeobox transcription factors across Metazoa and Archaeplastida.BMC Biol. 2020 Jan 9;18(1):4. doi: 10.1186/s12915-020-0737-2. BMC Biol. 2020. PMID: 31918709 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

