Transcriptomic and epigenetic profiling of 'diffuse midline gliomas, H3 K27M-mutant' discriminate two subgroups based on the type of histone H3 mutated and not supratentorial or infratentorial location
- PMID: 30396367
- PMCID: PMC6219253
- DOI: 10.1186/s40478-018-0614-1
Transcriptomic and epigenetic profiling of 'diffuse midline gliomas, H3 K27M-mutant' discriminate two subgroups based on the type of histone H3 mutated and not supratentorial or infratentorial location
Abstract
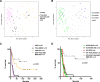
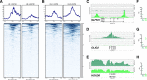
Diffuse midline glioma (DMG), H3 K27M-mutant, is a new entity in the updated WHO classification grouping together diffuse intrinsic pontine gliomas and infiltrating glial neoplasms of the midline harboring the same canonical mutation at the Lysine 27 of the histones H3 tail.Two hundred and fifteen patients younger than 18 years old with centrally-reviewed pediatric high-grade gliomas (pHGG) were included in this study. Comprehensive transcriptomic (n = 140) and methylation (n = 80) profiling was performed depending on the material available, in order to assess the biological uniqueness of this new entity compared to other midline and hemispheric pHGG.Tumor classification based on gene expression (GE) data highlighted the similarity of K27M DMG independently of their location along the midline. T-distributed Stochastic Neighbor Embedding (tSNE) analysis of methylation profiling confirms the discrimination of DMG from other well defined supratentorial tumor subgroups. Patients with diffuse intrinsic pontine gliomas (DIPG) and thalamic DMG exhibited a similarly poor prognosis (11.1 and 10.8 months median overall survival, respectively). Interestingly, H3.1-K27M and H3.3-K27M primary tumor samples could be distinguished based both on their GE and DNA methylation profiles, suggesting that they might arise from a different precursor or from a different epigenetic reorganization.These differences in DNA methylation profiles were conserved in glioma stem-like cell culture models of DIPG which mimicked their corresponding primary tumor. ChIP-seq profiling of H3K27me3 in these models indicate that H3.3-K27M mutated DIPG stem cells exhibit higher levels of H3K27 trimethylation which are correlated with fewer genes expressed by RNAseq. When considering the global distribution of the H3K27me3 mark, we observed that intergenic regions were more trimethylated in the H3.3-K27M mutated cells compared to the H3.1-K27M mutated ones.H3 K27M-mutant DMG represent a homogenous group of neoplasms compared to other pediatric gliomas that could be further separated based on the type of histone H3 variant mutated and their respective epigenetic landscapes. As these characteristics drive different phenotypes, these findings may have important implication for the design of future trials in these specific types of neoplasms.
Keywords: DNA methylation profiling; Diffuse intrinsic pontine glioma; Diffuse midline glioma; Epigenetics; Gene expression profiling; Glioma stem cell; H3 K27M-mutant; H3K27me3 landscape; Pediatric high-grade glioma.
Conflict of interest statement
Ethics approval and consent to participate
Informed consent for the translational research program was obtained from the parents or guardian according to the Institutional Review Board approved protocol (DC-2009- 955).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Castel D, Philippe C, Calmon R, Le Dret L, Truffaux N, Boddaert N, Pagès M, Taylor KR, Saulnier P, Lacroix L, Mackay A, Jones C, Sainte-Rose C, Blauwblomme T, Andreiuolo F, Puget S, Grill J, Varlet P, Debily M-A. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 2015;130:815–827. doi: 10.1007/s00401-015-1478-0. - DOI - PMC - PubMed
-
- Eisenstat DD, Pollack IF, Demers A, Sapp MV, Lambert P, Weisfeld-Adams JD, Burger PC, Gilles F, Davis RL, Packer R, Boyett JM, Finlay JL. Impact of tumor location and pathological discordance on survival of children with midline high-grade gliomas treated on Children’s Cancer Group high-grade glioma study CCG-945. J Neuro-Oncol. 2015;121:573–581. doi: 10.1007/s11060-014-1669-x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

