Genomic, Phenotypic, and Virulence Analysis of Streptococcus sanguinis Oral and Infective-Endocarditis Isolates
- PMID: 30396893
- PMCID: PMC6300630
- DOI: 10.1128/IAI.00703-18
Genomic, Phenotypic, and Virulence Analysis of Streptococcus sanguinis Oral and Infective-Endocarditis Isolates
Abstract
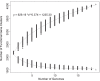
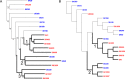
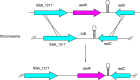
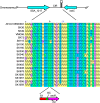
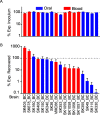
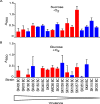
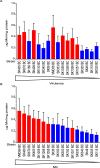
Streptococcus sanguinis, an abundant and benign inhabitant of the oral cavity, is an important etiologic agent of infective endocarditis (IE), particularly in people with predisposing cardiac valvular damage. Although commonly isolated from patients with IE, little is known about the factors that make any particular S. sanguinis isolate more virulent than another or, indeed, whether significant differences in virulence exist among isolates. In this study, we compared the genomes of a collection of S. sanguinis strains comprised of both oral isolates and bloodstream isolates from patients diagnosed with IE. Oral and IE isolates could not be distinguished by phylogenetic analyses, and we did not succeed in identifying virulence genes unique to the IE strains. We then investigated the virulence of these strains in a rabbit model of IE using a variation of the Bar-seq (barcode sequencing) method wherein we pooled the strains and used Illumina sequencing to count unique barcodes that had been inserted into each isolate at a conserved intergenic region. After we determined that several of the genome sequences were misidentified in GenBank, our virulence results were used to inform our bioinformatic analyses, identifying genes that may explain the heterogeneity in virulence. We further characterized these strains by assaying for phenotypes potentially contributing to virulence. Neither strain competition via bacteriocin production nor biofilm formation showed any apparent relationship with virulence. Increased cell-associated manganese was, however, correlated with blood isolates. These results, combined with additional phenotypic assays, suggest that S. sanguinis virulence is highly variable and results from multiple genetic factors.
Keywords: Streptococcus; biofilms; genomics; infective endocarditis; manganese; viridans; virulence.
Copyright © 2018 American Society for Microbiology.
Figures


Similar articles
-
Similar genomic patterns of clinical infective endocarditis and oral isolates of Streptococcus sanguinis and Streptococcus gordonii.Sci Rep. 2020 Feb 17;10(1):2728. doi: 10.1038/s41598-020-59549-4. Sci Rep. 2020. PMID: 32066773 Free PMC article.
-
Type IV Pili of Streptococcus sanguinis Contribute to Pathogenesis in Experimental Infective Endocarditis.Microbiol Spectr. 2021 Dec 22;9(3):e0175221. doi: 10.1128/Spectrum.01752-21. Epub 2021 Nov 10. Microbiol Spectr. 2021. PMID: 34756087 Free PMC article.
-
Identification of Streptococcus sanguinis genes required for biofilm formation and examination of their role in endocarditis virulence.Infect Immun. 2008 Jun;76(6):2551-9. doi: 10.1128/IAI.00338-08. Epub 2008 Apr 7. Infect Immun. 2008. PMID: 18390999 Free PMC article.
-
Streptococcus sanguinis biofilm formation & interaction with oral pathogens.Future Microbiol. 2018 Jun 1;13(8):915-932. doi: 10.2217/fmb-2018-0043. Epub 2018 Jun 8. Future Microbiol. 2018. PMID: 29882414 Free PMC article. Review.
-
Genomics, evolution, and molecular epidemiology of the Streptococcus bovis/Streptococcus equinus complex (SBSEC).Infect Genet Evol. 2015 Jul;33:419-36. doi: 10.1016/j.meegid.2014.09.017. Epub 2014 Sep 16. Infect Genet Evol. 2015. PMID: 25233845 Review.
Cited by
-
Shining Light on Oral Biofilm Fluorescence In Situ Hybridization (FISH): Probing the Accuracy of In Situ Biogeography Studies.Mol Oral Microbiol. 2025 Aug;40(4):137-146. doi: 10.1111/omi.12494. Epub 2025 Apr 30. Mol Oral Microbiol. 2025. PMID: 40304704
-
Comparative Genomics of Streptococcus oralis Identifies Large Scale Homologous Recombination and a Genetic Variant Associated with Infection.mSphere. 2022 Dec 21;7(6):e0050922. doi: 10.1128/msphere.00509-22. Epub 2022 Nov 2. mSphere. 2022. PMID: 36321824 Free PMC article.
-
Experimental evolution of gene essentiality in bacteria.bioRxiv [Preprint]. 2024 Dec 28:2024.07.16.600122. doi: 10.1101/2024.07.16.600122. bioRxiv. 2024. PMID: 39071448 Free PMC article. Preprint.
-
Intracellular Metal Speciation in Streptococcus sanguinis Establishes SsaACB as Critical for Redox Maintenance.ACS Infect Dis. 2020 Jul 10;6(7):1906-1921. doi: 10.1021/acsinfecdis.0c00132. Epub 2020 May 6. ACS Infect Dis. 2020. PMID: 32329608 Free PMC article.
-
Glycerol metabolism supports oral commensal interactions.ISME J. 2023 Jul;17(7):1116-1127. doi: 10.1038/s41396-023-01426-9. Epub 2023 May 11. ISME J. 2023. PMID: 37169870 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

