The multitasking polyA tail: nuclear RNA maturation, degradation and export
- PMID: 30397105
- PMCID: PMC6232593
- DOI: 10.1098/rstb.2018.0169
The multitasking polyA tail: nuclear RNA maturation, degradation and export
Abstract
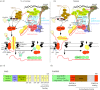
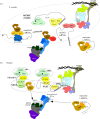
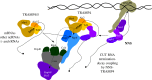
A polyA (pA) tail is an essential modification added to the 3' ends of a wide range of RNAs at different stages of their metabolism. Here, we describe the main sources of polyadenylation and outline their underlying biochemical interactions within the nuclei of budding yeast Saccharomyces cerevisiae, human cells and, when relevant, the fission yeast Schizosaccharomyces pombe Polyadenylation mediated by the S. cerevisiae Trf4/5 enzymes, and their human homologues PAPD5/7, typically leads to the 3'-end trimming or complete decay of non-coding RNAs. By contrast, the primary function of canonical pA polymerases (PAPs) is to produce stable and nuclear export-competent mRNAs. However, this dichotomy is becoming increasingly blurred, at least in S. pombe and human cells, where polyadenylation mediated by canonical PAPs may also result in transcript decay.This article is part of the theme issue '5' and 3' modifications controlling RNA degradation'.
Keywords: RNA decay; RNA export; RNA polyadenylation; TRAMP complex; polyA binding proteins; transcription termination.
© 2018 The Author(s).
Conflict of interest statement
We declare we have no competing interests.
Figures
Similar articles
-
Requirement of fission yeast Cid14 in polyadenylation of rRNAs.Mol Cell Biol. 2006 Mar;26(5):1710-21. doi: 10.1128/MCB.26.5.1710-1721.2006. Mol Cell Biol. 2006. PMID: 16478992 Free PMC article.
-
The Nuclear RNA Exosome and Its Cofactors.Adv Exp Med Biol. 2019;1203:113-132. doi: 10.1007/978-3-030-31434-7_4. Adv Exp Med Biol. 2019. PMID: 31811632 Review.
-
The Nrd1-like protein Seb1 coordinates cotranscriptional 3' end processing and polyadenylation site selection.Genes Dev. 2016 Jul 1;30(13):1558-72. doi: 10.1101/gad.280222.116. Genes Dev. 2016. PMID: 27401558 Free PMC article.
-
Comparative analysis of alternative polyadenylation in S. cerevisiae and S. pombe.Genome Res. 2017 Oct;27(10):1685-1695. doi: 10.1101/gr.222331.117. Epub 2017 Sep 15. Genome Res. 2017. PMID: 28916539 Free PMC article.
-
The nuclear RNA surveillance machinery: the link between ncRNAs and genome structure in budding yeast?Biochim Biophys Acta. 2008 Apr;1779(4):239-46. doi: 10.1016/j.bbagrm.2007.12.008. Epub 2008 Jan 3. Biochim Biophys Acta. 2008. PMID: 18211833 Review.
Cited by
-
The RNA export and RNA decay complexes THO and TRAMP prevent transcription-replication conflicts, DNA breaks, and CAG repeat contractions.PLoS Biol. 2022 Dec 27;20(12):e3001940. doi: 10.1371/journal.pbio.3001940. eCollection 2022 Dec. PLoS Biol. 2022. PMID: 36574440 Free PMC article.
-
System-wide analyses of the fission yeast poly(A)+ RNA interactome reveal insights into organization and function of RNA-protein complexes.Genome Res. 2020 Jul;30(7):1012-1026. doi: 10.1101/gr.257006.119. Epub 2020 Jun 18. Genome Res. 2020. PMID: 32554781 Free PMC article.
-
Recent molecular insights into canonical pre-mRNA 3'-end processing.Transcription. 2020 Apr;11(2):83-96. doi: 10.1080/21541264.2020.1777047. Epub 2020 Jun 11. Transcription. 2020. PMID: 32522085 Free PMC article. Review.
-
Altered rRNA processing disrupts nuclear RNA homeostasis via competition for the poly(A)-binding protein Nab2.Nucleic Acids Res. 2020 Nov 18;48(20):11675-11694. doi: 10.1093/nar/gkaa964. Nucleic Acids Res. 2020. PMID: 33137177 Free PMC article.
-
The Post-Transcriptional Regulatory Element of Hepatitis B Virus: From Discovery to Therapy.Viruses. 2024 Mar 29;16(4):528. doi: 10.3390/v16040528. Viruses. 2024. PMID: 38675871 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

