Crystal structure of heme A synthase from Bacillus subtilis
- PMID: 30397130
- PMCID: PMC6255202
- DOI: 10.1073/pnas.1813346115
Crystal structure of heme A synthase from Bacillus subtilis
Abstract
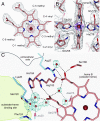
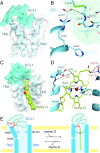
Heme A is an essential cofactor for respiratory terminal oxidases and vital for respiration in aerobic organisms. The final step of heme A biosynthesis is formylation of the C-8 methyl group of heme molecule by heme A synthase (HAS). HAS is a heme-containing integral membrane protein, and its structure and reaction mechanisms have remained unknown. Thus, little is known about HAS despite of its importance. Here we report the crystal structure of HAS from Bacillus subtilis at 2.2-Å resolution. The N- and C-terminal halves of HAS consist of four-helix bundles and they align in a pseudo twofold symmetry manner. Each bundle contains a pair of histidine residues and forms a heme-binding domain. The C-half domain binds a cofactor-heme molecule, while the N-half domain is vacant. Many water molecules are found in the transmembrane region and around the substrate-binding site, and some of them interact with the main chain of transmembrane helix. Comparison of these two domain structures enables us to construct a substrate-heme binding state structure. This structure implies that a completely conserved glutamate, Glu57 in B. subtilis, is the catalytic residue for the formylation reaction. These results provide valuable suggestions of the substrate-heme binding mechanism. Our results present significant insight into the heme A biosynthesis.
Keywords: CtaA; crystal structure; formylation reaction; heme A biosynthesis; membrane protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Tsukihara T, et al. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 A. Science. 1995;269:1069–1074. - PubMed
-
- Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. - PubMed
-
- Yoshikawa S, Shimada A. Reaction mechanism of cytochrome c oxidase. Chem Rev. 2015;115:1936–1989. - PubMed
-
- Saiki K, Mogi T, Anraku Y. Heme O biosynthesis in Escherichia coli: The cyoE gene in the cytochrome bo operon encodes a protoheme IX farnesyltransferase. Biochem Biophys Res Commun. 1992;189:1491–1497. - PubMed
-
- Svensson B, Lübben M, Hederstedt L. Bacillus subtilis CtaA and CtaB function in haem A biosynthesis. Mol Microbiol. 1993;10:193–201. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

