Expanded skin virome in DOCK8-deficient patients
- PMID: 30397357
- PMCID: PMC6286253
- DOI: 10.1038/s41591-018-0211-7
Expanded skin virome in DOCK8-deficient patients
Abstract
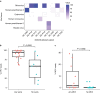
Human microbiome studies have revealed the intricate interplay of host immunity and bacterial communities to achieve homeostatic balance. Healthy skin microbial communities are dominated by bacteria with low viral representation1-3, mainly bacteriophage. Specific eukaryotic viruses have been implicated in both common and rare skin diseases, but cataloging skin viral communities has been limited. Alterations in host immunity provide an opportunity to expand our understanding of microbial-host interactions. Primary immunodeficient patients manifest with various viral, bacterial, fungal, and parasitic infections, including skin infections4. Dedicator of cytokinesis 8 (DOCK8) deficiency is a rare primary human immunodeficiency characterized by recurrent cutaneous and systemic infections, as well as atopy and cancer susceptibility5. DOCK8, encoding a guanine nucleotide exchange factor highly expressed in lymphocytes, regulates actin cytoskeleton, which is critical for migration through collagen-dense tissues such as skin6. Analyzing deep metagenomic sequencing data from DOCK8-deficient skin samples demonstrated a notable increase in eukaryotic viral representation and diversity compared with healthy volunteers. De novo assembly approaches identified hundreds of novel human papillomavirus genomes, illuminating microbial dark matter. Expansion of the skin virome in DOCK8-deficient patients underscores the importance of immune surveillance in controlling eukaryotic viral colonization and infection.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

