Methylation divergence of invasive Ciona ascidians: Significant population structure and local environmental influence
- PMID: 30397465
- PMCID: PMC6206186
- DOI: 10.1002/ece3.4504
Methylation divergence of invasive Ciona ascidians: Significant population structure and local environmental influence
Abstract
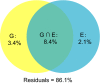
The geographical expansion of invasive species usually leads to temporary and/or permanent changes at multiple levels (genetics, epigenetics, gene expression, etc.) to acclimatize to abiotic and/or biotic stresses in novel environments. Epigenetic variation such as DNA methylation is often involved in response to diverse local environments, thus representing one crucial mechanism to promote invasion success. However, evidence is scant on the potential role of DNA methylation variation in rapid environmental response and invasion success during biological invasions. In particular, DNA methylation patterns and possible contributions of varied environmental factors to methylation differentiation have been largely unknown in many invaders, especially for invasive species in marine systems where extremely complex interactions exist between species and surrounding environments. Using the methylation-sensitive amplification polymorphism (MSAP) technique, here we investigated population methylation structure at the genome level in two highly invasive model ascidians, Ciona robusta and C. intestinalis, collected from habitats with varied environmental factors such as temperature and salinity. We found high intrapopulation methylation diversity and significant population methylation differentiation in both species. Multiple analyses, such as variation partitioning analysis, showed that both genetic variation and environmental factors contributed to the observed DNA methylation variation. Further analyses found that 24 and 20 subepiloci were associated with temperature and/or salinity in C. robusta and C. intestinalis, respectively. All these results clearly showed significant methylation divergence among populations of both invasive ascidians, and varied local environmental factors, as well as genetic variation, were responsible for the observed DNA methylation patterns. The consistent findings in both species here suggest that DNA methylation, coupled with genetic variation, may facilitate local environmental adaptation during biological invasions, and DNA methylation variation molded by local environments may contribute to invasion success.
Keywords: DNA methylation; biological invasion; methylation divergence; methylation‐sensitive amplified polymorphism; tunicate.
Figures





References
Associated data
LinkOut - more resources
Full Text Sources

