Hidden diversity and evolution of viruses in market fish
- PMID: 30397510
- PMCID: PMC6208713
- DOI: 10.1093/ve/vey031
Hidden diversity and evolution of viruses in market fish
Abstract
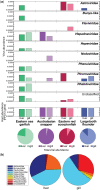
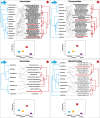
Aquaculture is the fastest growing industry worldwide. Aquatic diseases have had enormous economic and environmental impacts in the recent past and the emergence of new aquatic pathogens, particularly viruses, poses a continuous threat. Nevertheless, little is known about the diversity, abundance and evolution of fish viruses. We used a meta-transcriptomic approach to help determine the virome of seemingly healthy fish sold at a market in Sydney, Australia. Specifically, by identifying and quantifying virus transcripts we aimed to determine (i) the abundance of viruses in market fish, (ii) test a key component of epidemiological theory that large and dense host populations harbour a greater number of viruses compared to their more solitary counterparts and (iii) reveal the relative roles of virus-host co-divergence and cross-species transmission in the evolution of fish viruses. The species studied comprised both shoaling fish-eastern sea garfish (Hyporhamphus australis) and Australasian snapper (Chrysophrys auratus)-and more solitary fish-eastern red scorpionfish (Scorpaena jacksoniensis) and largetooth flounder (Pseudorhombus arsius). Our analysis identified twelve potentially novel viruses, eight of which were likely vertebrate-associated across four viral families and that exhibited frequent cross-species transmission. Notably, the most solitary of the fish species studied, the largetooth flounder, harboured the least number of viruses while eastern sea garfish, a densely shoaling fish, had the highest number of viruses. These results support the emerging view that fish harbour a large and largely uncharacterised virome.
Keywords: fish; meta-transcriptomics; phylogenetics; virome; virus evolution.
Figures



References
-
- Anderson R. M., May R. M. (1982) ‘Coevolution of Hosts and Parasites’, Parasitology, 85: 411–26. - PubMed
-
- Barbknecht M. et al. (2014) ‘Characterization of a New Picornavirus Isolated from the Freshwater Fish Lepomis Macrochirus’, Journal of General Virology, 95: 601–13. - PubMed
-
- Broitman B. R. et al. (2017) ‘Dynamic Interactions among Boundaries and the Expansion of Sustainable Aquaculture’, Frontiers in Marine Science, 4: 1–15.
LinkOut - more resources
Full Text Sources
Miscellaneous

