Uncovering full-length transcript isoforms of sugarcane cultivar Khon Kaen 3 using single-molecule long-read sequencing
- PMID: 30397543
- PMCID: PMC6214230
- DOI: 10.7717/peerj.5818
Uncovering full-length transcript isoforms of sugarcane cultivar Khon Kaen 3 using single-molecule long-read sequencing
Abstract
Background: Sugarcane is an important global food crop and energy resource. To facilitate the sugarcane improvement program, genome and gene information are important for studying traits at the molecular level. Most currently available transcriptome data for sugarcane were generated using second-generation sequencing platforms, which provide short reads. The de novo assembled transcripts from these data are limited in length, and hence may be incomplete and inaccurate, especially for long RNAs.
Methods: We generated a transcriptome dataset of leaf tissue from a commercial Thai sugarcane cultivar Khon Kaen 3 (KK3) using PacBio RS II single-molecule long-read sequencing by the Iso-Seq method. Short-read RNA-Seq data were generated from the same RNA sample using the Ion Proton platform for reducing base calling errors.
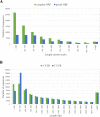
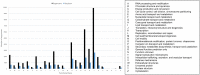
Results: A total of 119,339 error-corrected transcripts were generated with the N50 length of 3,611 bp, which is on average longer than any previously reported sugarcane transcriptome dataset. 110,253 sequences (92.4%) contain an open reading frame (ORF) of at least 300 bp long with ORF N50 of 1,416 bp. The mean lengths of 5' and 3' untranslated regions in 73,795 sequences with complete ORFs are 1,249 and 1,187 bp, respectively. 4,774 transcripts are putatively novel full-length transcripts which do not match with a previous Iso-Seq study of sugarcane. We annotated the functions of 68,962 putative full-length transcripts with at least 90% coverage when compared with homologous protein coding sequences in other plants.
Discussion: The new catalog of transcripts will be useful for genome annotation, identification of splicing variants, SNP identification, and other research pertaining to the sugarcane improvement program. The putatively novel transcripts suggest unique features of KK3, although more data from different tissues and stages of development are needed to establish a reference transcriptome of this cultivar.
Keywords: Full-length transcripts; Iso-Seq; KK3; Khon Kaen 3; PacBio sequencing; Single-molecule long-read sequencing; Sugarcane; Transcriptome.
Conflict of interest statement
Warodom Wirojsirasak, Prapat Punpee and Peeraya Klomsa-ard are employed by Mitr Phol Sugarcane Research Center Co., Ltd.
Figures





Similar articles
-
A survey of the complex transcriptome from the highly polyploid sugarcane genome using full-length isoform sequencing and de novo assembly from short read sequencing.BMC Genomics. 2017 May 22;18(1):395. doi: 10.1186/s12864-017-3757-8. BMC Genomics. 2017. PMID: 28532419 Free PMC article.
-
A de novo Full-Length mRNA Transcriptome Generated From Hybrid-Corrected PacBio Long-Reads Improves the Transcript Annotation and Identifies Thousands of Novel Splice Variants in Atlantic Salmon.Front Genet. 2021 Apr 27;12:656334. doi: 10.3389/fgene.2021.656334. eCollection 2021. Front Genet. 2021. PMID: 33986770 Free PMC article.
-
Unveiling the transcriptomic complexity of Miscanthus sinensis using a combination of PacBio long read- and Illumina short read sequencing platforms.BMC Genomics. 2021 Sep 22;22(1):690. doi: 10.1186/s12864-021-07971-x. BMC Genomics. 2021. PMID: 34551715 Free PMC article.
-
Methodologies for Transcript Profiling Using Long-Read Technologies.Front Genet. 2020 Jul 7;11:606. doi: 10.3389/fgene.2020.00606. eCollection 2020. Front Genet. 2020. PMID: 32733532 Free PMC article. Review.
-
Reviving the Transcriptome Studies: An Insight Into the Emergence of Single-Molecule Transcriptome Sequencing.Front Genet. 2019 Apr 26;10:384. doi: 10.3389/fgene.2019.00384. eCollection 2019. Front Genet. 2019. PMID: 31105749 Free PMC article. Review.
Cited by
-
Taxonomically Restricted Genes Are Associated With Responses to Biotic and Abiotic Stresses in Sugarcane (Saccharum spp.).Front Plant Sci. 2022 Jun 30;13:923069. doi: 10.3389/fpls.2022.923069. eCollection 2022. Front Plant Sci. 2022. PMID: 35845637 Free PMC article.
-
Characterization of full-length transcriptome in Saccharum officinarum and molecular insights into tiller development.BMC Plant Biol. 2021 May 22;21(1):228. doi: 10.1186/s12870-021-02989-5. BMC Plant Biol. 2021. PMID: 34022806 Free PMC article.
-
A high-resolution single-molecule sequencing-based Arabidopsis transcriptome using novel methods of Iso-seq analysis.Genome Biol. 2022 Jul 7;23(1):149. doi: 10.1186/s13059-022-02711-0. Genome Biol. 2022. PMID: 35799267 Free PMC article.
-
Amino Acid and Carbohydrate Metabolism Are Coordinated to Maintain Energetic Balance during Drought in Sugarcane.Int J Mol Sci. 2020 Nov 30;21(23):9124. doi: 10.3390/ijms21239124. Int J Mol Sci. 2020. PMID: 33266228 Free PMC article.
-
A SNP variation in the Sucrose synthase (SoSUS) gene associated with sugar-related traits in sugarcane.PeerJ. 2023 Dec 15;11:e16667. doi: 10.7717/peerj.16667. eCollection 2023. PeerJ. 2023. PMID: 38111652 Free PMC article.
References
-
- Au KF, Sebastiano V, Afshar PT, Durruthy JD, Lee L, Williams BA, Van Bakel H, Schadt EE, Reijo-Pera RA, Underwood JG, Wong WH. Characterization of the human ESC transcriptome by hybrid sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E4821–E4830. doi: 10.1073/pnas.1320101110. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

