Structure and dynamics conspire in the evolution of affinity between intrinsically disordered proteins
- PMID: 30397651
- PMCID: PMC6200366
- DOI: 10.1126/sciadv.aau4130
Structure and dynamics conspire in the evolution of affinity between intrinsically disordered proteins
Abstract
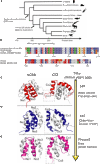
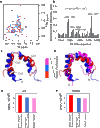
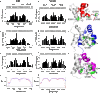
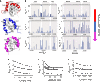
In every established species, protein-protein interactions have evolved such that they are fit for purpose. However, the molecular details of the evolution of new protein-protein interactions are poorly understood. We have used nuclear magnetic resonance spectroscopy to investigate the changes in structure and dynamics during the evolution of a protein-protein interaction involving the intrinsically disordered CREBBP (CREB-binding protein) interaction domain (CID) and nuclear coactivator binding domain (NCBD) from the transcriptional coregulators NCOA (nuclear receptor coactivator) and CREBBP/p300, respectively. The most ancient low-affinity "Cambrian-like" [540 to 600 million years (Ma) ago] CID/NCBD complex contained less secondary structure and was more dynamic than the complexes from an evolutionarily younger "Ordovician-Silurian" fish ancestor (ca. 440 Ma ago) and extant human. The most ancient Cambrian-like CID/NCBD complex lacked one helix and several interdomain interactions, resulting in a larger solvent-accessible surface area. Furthermore, the most ancient complex had a high degree of millisecond-to-microsecond dynamics distributed along the entire sequences of both CID and NCBD. These motions were reduced in the Ordovician-Silurian CID/NCBD complex and further redistributed in the extant human CID/NCBD complex. Isothermal calorimetry experiments show that complex formation is enthalpically favorable and that affinity is modulated by a largely unfavorable entropic contribution to binding. Our data demonstrate how changes in structure and motion conspire to shape affinity during the evolution of a protein-protein complex and provide direct evidence for the role of structural, dynamic, and frustrational plasticity in the evolution of interactions between intrinsically disordered proteins.
Figures
Similar articles
-
αα-Hub domains and intrinsically disordered proteins: A decisive combo.J Biol Chem. 2021 Jan-Jun;296:100226. doi: 10.1074/jbc.REV120.012928. Epub 2020 Dec 29. J Biol Chem. 2021. PMID: 33361159 Free PMC article. Review.
-
Mapping the transition state for a binding reaction between ancient intrinsically disordered proteins.J Biol Chem. 2020 Dec 18;295(51):17698-17712. doi: 10.1074/jbc.RA120.015645. J Biol Chem. 2020. PMID: 33454008 Free PMC article.
-
Emergence and evolution of an interaction between intrinsically disordered proteins.Elife. 2017 Apr 11;6:e16059. doi: 10.7554/eLife.16059. Elife. 2017. PMID: 28398197 Free PMC article.
-
The dynamic properties of a nuclear coactivator binding domain are evolutionarily conserved.Commun Biol. 2022 Mar 30;5(1):286. doi: 10.1038/s42003-022-03217-y. Commun Biol. 2022. PMID: 35354917 Free PMC article.
-
Coupled binding and folding of intrinsically disordered proteins: what can we learn from kinetics?Curr Opin Struct Biol. 2016 Feb;36:18-24. doi: 10.1016/j.sbi.2015.11.012. Epub 2015 Dec 22. Curr Opin Struct Biol. 2016. PMID: 26720267 Review.
Cited by
-
IFF: Identifying key residues in intrinsically disordered regions of proteins using machine learning.Protein Sci. 2023 Sep;32(9):e4739. doi: 10.1002/pro.4739. Protein Sci. 2023. PMID: 37498545 Free PMC article.
-
Structural and Dynamical Order of a Disordered Protein: Molecular Insights into Conformational Switching of PAGE4 at the Systems Level.Biomolecules. 2019 Feb 22;9(2):77. doi: 10.3390/biom9020077. Biomolecules. 2019. PMID: 30813315 Free PMC article.
-
Ancestral sequence reconstruction as a tool to study the evolution of wood decaying fungi.Front Fungal Biol. 2022 Oct 14;3:1003489. doi: 10.3389/ffunb.2022.1003489. eCollection 2022. Front Fungal Biol. 2022. PMID: 37746217 Free PMC article. Review.
-
A billion years of evolution manifest in nanosecond protein dynamics.Proc Natl Acad Sci U S A. 2024 Mar 5;121(10):e2318743121. doi: 10.1073/pnas.2318743121. Epub 2024 Feb 27. Proc Natl Acad Sci U S A. 2024. PMID: 38412135 Free PMC article.
-
αα-Hub domains and intrinsically disordered proteins: A decisive combo.J Biol Chem. 2021 Jan-Jun;296:100226. doi: 10.1074/jbc.REV120.012928. Epub 2020 Dec 29. J Biol Chem. 2021. PMID: 33361159 Free PMC article. Review.
References
-
- Leo C., Chen J. D., The SRC family of nuclear receptor coactivators. Gene 245, 1–11 (2000). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

