Dominant-negative SERPING1 variants cause intracellular retention of C1 inhibitor in hereditary angioedema
- PMID: 30398465
- PMCID: PMC6307969
- DOI: 10.1172/JCI98869
Dominant-negative SERPING1 variants cause intracellular retention of C1 inhibitor in hereditary angioedema
Abstract
Hereditary angioedema (HAE) is an autosomal dominant disease characterized by recurrent edema attacks associated with morbidity and mortality. HAE results from variations in the SERPING1 gene that encodes the C1 inhibitor (C1INH), a serine protease inhibitor (serpin). Reduced plasma levels of C1INH lead to enhanced activation of the contact system, triggering high levels of bradykinin and increased vascular permeability, but the cellular mechanisms leading to low C1INH levels (20%-30% of normal) in heterozygous HAE type I patients remain obscure. Here, we showed that C1INH encoded by a subset of HAE-causing SERPING1 alleles affected secretion of normal C1INH protein in a dominant-negative fashion by triggering formation of protein-protein interactions between normal and mutant C1INH, leading to the creation of larger intracellular C1INH aggregates that were trapped in the endoplasmic reticulum (ER). Notably, intracellular aggregation of C1INH and ER abnormality were observed in fibroblasts from a heterozygous carrier of a dominant-negative SERPING1 gene variant, but the condition was ameliorated by viral delivery of the SERPING1 gene. Collectively, our data link abnormal accumulation of serpins, a hallmark of serpinopathies, with dominant-negative disease mechanisms affecting C1INH plasma levels in HAE type I patients, and may pave the way for new treatments of HAE.
Keywords: Cell Biology; Genetic diseases; Genetics; Molecular genetics; Serpins.
Conflict of interest statement
Figures



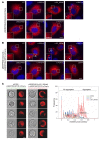
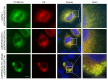

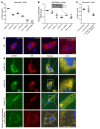



Comment in
-
The hereditary angioedema syndromes.J Clin Invest. 2019 Jan 2;129(1):66-68. doi: 10.1172/JCI125378. Epub 2018 Dec 10. J Clin Invest. 2019. PMID: 30530986 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

