InterPro in 2019: improving coverage, classification and access to protein sequence annotations
- PMID: 30398656
- PMCID: PMC6323941
- DOI: 10.1093/nar/gky1100
InterPro in 2019: improving coverage, classification and access to protein sequence annotations
Abstract
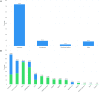
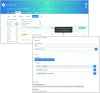
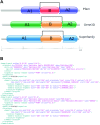
The InterPro database (http://www.ebi.ac.uk/interpro/) classifies protein sequences into families and predicts the presence of functionally important domains and sites. Here, we report recent developments with InterPro (version 70.0) and its associated software, including an 18% growth in the size of the database in terms on new InterPro entries, updates to content, the inclusion of an additional entry type, refined modelling of discontinuous domains, and the development of a new programmatic interface and website. These developments extend and enrich the information provided by InterPro, and provide greater flexibility in terms of data access. We also show that InterPro's sequence coverage has kept pace with the growth of UniProtKB, and discuss how our evaluation of residue coverage may help guide future curation activities.
Figures




References
Publication types
MeSH terms
Grants and funding
- U41 HG002273/HG/NHGRI NIH HHS/United States
- BB/N019172/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/L024136/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/N00521X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

