Multidimensional Assessment of the Host Response in Mechanically Ventilated Patients with Suspected Pneumonia
- PMID: 30398927
- PMCID: PMC6519857
- DOI: 10.1164/rccm.201804-0650OC
Multidimensional Assessment of the Host Response in Mechanically Ventilated Patients with Suspected Pneumonia
Abstract
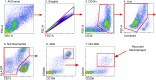
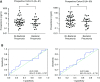
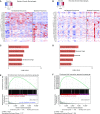
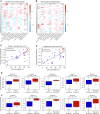
Rationale: The identification of informative elements of the host response to infection may improve the diagnosis and management of bacterial pneumonia. Objectives: To determine whether the absence of alveolar neutrophilia can exclude bacterial pneumonia in critically ill patients with suspected infection and to test whether signatures of bacterial pneumonia can be identified in the alveolar macrophage transcriptome. Methods: We determined the test characteristics of alveolar neutrophilia for the diagnosis of bacterial pneumonia in three cohorts of mechanically ventilated patients. In one cohort, we also isolated macrophages from alveolar lavage fluid and used the transcriptome to identify signatures of bacterial pneumonia. Finally, we developed a humanized mouse model of Pseudomonas aeruginosa pneumonia to determine if pathogen-specific signatures can be identified in human alveolar macrophages. Measurements and Main Results: An alveolar neutrophil percentage less than 50% had a negative predictive value of greater than 90% for bacterial pneumonia in both the retrospective (n = 851) and validation cohorts (n = 76 and n = 79). A transcriptional signature of bacterial pneumonia was present in both resident and recruited macrophages. Gene signatures from both cell types identified patients with bacterial pneumonia with test characteristics similar to alveolar neutrophilia. Conclusions: The absence of alveolar neutrophilia has a high negative predictive value for bacterial pneumonia in critically ill patients with suspected infection. Macrophages can be isolated from alveolar lavage fluid obtained during routine care and used for RNA-Seq analysis. This novel approach may facilitate a longitudinal and multidimensional assessment of the host response to bacterial pneumonia.
Keywords: RNA-Seq; alveolar macrophages; bacterial pneumonia; host response.
Figures

Comment in
-
Teaching an Old Intensivist Neutrophil Tricks: Using Alveolar Neutrophilia to Diagnose Ventilator-associated Pneumonia.Am J Respir Crit Care Med. 2019 May 15;199(10):1178-1181. doi: 10.1164/rccm.201811-2163ED. Am J Respir Crit Care Med. 2019. PMID: 30543457 Free PMC article. No abstract available.
References
-
- Flanders SA, Saint S. Why does antimicrobial overuse in hospitalized patients persist? JAMA Intern Med. 2014;174:661–662. - PubMed
-
- Marston HD, Dixon DM, Knisely JM, Palmore TN, Fauci AS. Antimicrobial resistance. JAMA. 2016;316:1193–1204. - PubMed
-
- Shindo Y, Ito R, Kobayashi D, Ando M, Ichikawa M, Shiraki A, et al. Risk factors for drug-resistant pathogens in community-acquired and healthcare-associated pneumonia. Am J Respir Crit Care Med. 2013;188:985–995. - PubMed
-
- Xu J, Murphy SL, Kochanek KD, Bastian BA. Deaths: final data for 2013. Natl Vital Stat Rep. 2016;64:1–119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- I01 CX001777/CX/CSRD VA/United States
- R01 ES013995/ES/NIEHS NIH HHS/United States
- K08 HL125940/HL/NHLBI NIH HHS/United States
- I01 BX000201/BX/BLRD VA/United States
- P01 AG049665/AG/NIA NIH HHS/United States
- P01 HL071643/HL/NHLBI NIH HHS/United States
- U19 AI135964/AI/NIAID NIH HHS/United States
- UL1 TR001422/TR/NCATS NIH HHS/United States
- P30 CA060553/CA/NCI NIH HHS/United States
- T32 HL076139/HL/NHLBI NIH HHS/United States
- S10 OD011996/OD/NIH HHS/United States
- R03 AR061593/AR/NIAMS NIH HHS/United States
- F32 HL136111/HL/NHLBI NIH HHS/United States
- R56 HL135124/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

