Detection of High-Risk Human Papillomavirus in Oral Cavity Squamous Cell Carcinoma Using Multiple Analytes and Their Role in Patient Survival
- PMID: 30398949
- PMCID: PMC7010445
- DOI: 10.1200/JGO.18.00058
Detection of High-Risk Human Papillomavirus in Oral Cavity Squamous Cell Carcinoma Using Multiple Analytes and Their Role in Patient Survival
Abstract
Purpose: Accurate detection of human papillomavirus (HPV) in oral cavity squamous cell carcinoma (OSCC) is essential to understanding the role of HPV in disease prognosis and management of patients. We used different analytes and methods to understand the true prevalence of HPV in a cohort of patients with OSCC with different molecular backgrounds, and we correlated HPV data with patient survival.
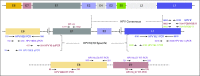
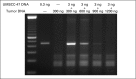
Methods: We integrated data from multiple analytes (HPV DNA, HPV RNA, and p16), assays (immunohistochemistry, polymerase chain reaction [PCR], quantitative PCR [qPCR], and digital PCR), and molecular changes (somatic mutations and DNA methylation) from 153 patients with OSCC to correlate p16 expression, HPV DNA, and HPV RNA with HPV incidence and patient survival.
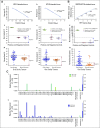
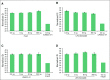
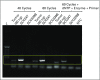
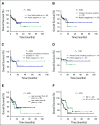
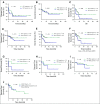
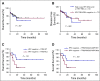
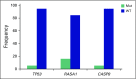
Results: High prevalence (33% to 58%) of HPV16/18 DNA did not correlate with the presence of transcriptionally active viral genomes (15%) in tumors. Eighteen percent of the tumors were p16 positive and only 6% were both HPV DNA and HPV RNA positive. Most tumors with relatively high copy number HPV DNA and/or HPV RNA, but not with HPV DNA alone (irrespective of copy number), were wild-type for TP53 and CASP8 genes. In our study, p16 protein, HPV DNA, and HPV RNA, either alone or in combination, did not correlate with patient survival. Nine HPV-associated genes stratified the virus-positive from the virus-negative tumor group with high confidence ( P < .008) when HPV DNA copy number and/or HPV RNA were considered to define HPV positivity, and not HPV DNA alone, irrespective of copy number ( P < .2).
Conclusion: In OSCC, the presence of both HPV RNA and p16 is rare. HPV DNA alone is not an accurate measure of HPV positivity and therefore may not be informative. HPV DNA, HPV RNA, and p16 do not correlate with patients' outcome.
Conflict of interest statement
Vinayak Palve
No relationship to disclose
Jamir Bagwan
No relationship to disclose
Neeraja M. Krishnan
No relationship to disclose
Manisha Pareek
No relationship to disclose
Udita Chandola
No relationship to disclose
Amritha Suresh
No relationship to disclose
Gangotri Siddappa
No relationship to disclose
Bonney L. James
No relationship to disclose
Vikram Kekatpure
No relationship to disclose
Moni Abraham Kuriakose
No relationship to disclose
Binay Panda
No relationship to disclose
Figures





Similar articles
-
Detection of human papillomavirus infection in oral squamous cell carcinoma: a cohort study of Japanese patients.J Oral Pathol Med. 2016 Sep;45(8):565-72. doi: 10.1111/jop.12416. Epub 2015 Dec 29. J Oral Pathol Med. 2016. PMID: 26711722
-
Lack of evidence of human papillomavirus-induced squamous cell carcinomas of the oral cavity in southern Germany.Oral Oncol. 2013 Sep;49(9):937-942. doi: 10.1016/j.oraloncology.2013.03.451. Epub 2013 Apr 19. Oral Oncol. 2013. PMID: 23608471
-
The role of human papillomavirus in p16-positive oral cancers.J Oral Pathol Med. 2018 Jan;47(1):18-24. doi: 10.1111/jop.12649. Epub 2017 Oct 30. J Oral Pathol Med. 2018. PMID: 29024035
-
HPV and cancer of the oral cavity.Virulence. 2015;6(3):244-8. doi: 10.1080/21505594.2014.999570. Virulence. 2015. PMID: 25654476 Free PMC article. Review.
-
[Oral squamous cell carcinoma. Case report and review of literature].Rev Med Inst Mex Seguro Soc. 2022 Feb 1;60(1):85-90. Rev Med Inst Mex Seguro Soc. 2022. PMID: 35274916 Free PMC article. Review. Spanish.
Cited by
-
Demographic Profile of p16 Immunopositive and HPV DNA PCR Positive Oral Squamous Cell Carcinoma in a Large Cohort of Indian Patients.Asian Pac J Cancer Prev. 2022 Feb 1;23(2):529-536. doi: 10.31557/APJCP.2022.23.2.529. Asian Pac J Cancer Prev. 2022. PMID: 35225465 Free PMC article.
-
The Prevalence of HPV in Oral Cavity Squamous Cell Carcinoma.Viruses. 2023 Feb 6;15(2):451. doi: 10.3390/v15020451. Viruses. 2023. PMID: 36851665 Free PMC article. Review.
-
Concordance of p16INK4a and E6*I mRNA among HPV-DNA-Positive Oropharyngeal, Laryngeal, and Oral Cavity Carcinomas from the ICO International Study.Cancers (Basel). 2022 Aug 4;14(15):3787. doi: 10.3390/cancers14153787. Cancers (Basel). 2022. PMID: 35954451 Free PMC article.
-
HPV in non-oropharyngeal head and neck cancer: does it matter?Ann Transl Med. 2020 Sep;8(18):1120. doi: 10.21037/atm-20-3346. Ann Transl Med. 2020. PMID: 33240969 Free PMC article. No abstract available.
-
The correlation between human papillomavirus and oral lichen planus: A systematic review of the literature.Immun Inflamm Dis. 2023 Aug;11(8):e960. doi: 10.1002/iid3.960. Immun Inflamm Dis. 2023. PMID: 37647448 Free PMC article.
References
-
- Stenson KM, Brockstein BE, Ross ME. Epidemiology and risk factors for head and neck cancer. 2014 https://www.uptodate.com/contents/epidemiology-and-risk-factors-for-head...
-
- Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. - PubMed
-
- Bhat SP, Bhat V, Permi H, et al. Oral and oropharyngeal malignancy: A clinicopathological study. Internet J Pathol Lab Med. 2016;2:OA3. https://www.chanrejournals.com/index.php/pathology/article/view/129/html
-
- Mishra A, Meherotra R. Head and neck cancer: Global burden and regional trends in India. Asian Pac J Cancer Prev. 2014;15:537–550. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

