Transcriptome Analysis of Gene Expression Patterns Potentially Associated with Premature Senescence in Nicotiana tabacum L
- PMID: 30400189
- PMCID: PMC6278766
- DOI: 10.3390/molecules23112856
Transcriptome Analysis of Gene Expression Patterns Potentially Associated with Premature Senescence in Nicotiana tabacum L
Abstract
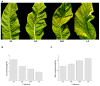
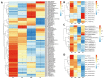
Senescence affects the remobilization of nutrients and adaption of the plant to the environment. Combined stresses can result in premature senescence in plants which exist in the field. In this study, transcriptomic analysis was performed on mature leaves and leaves in three stages of premature senescence to understand the molecular mechanism. With progressive premature senescence, a declining chlorophyll (chl) content and an increasing malonaldehyde (MDA) content were observed, while plasmolysis and cell nucleus pyknosis occurred, mitochondria melted, thylakoid lamellae were dilated, starch grains in chloroplast decreased, and osmiophilic granules increased gradually. Moreover, in total 69 common differentially expressed genes (DEGs) in three stages of premature senescing leaves were found, which were significantly enriched in summarized Gene Ontology (GO) terms of membrane-bounded organelle, regulation of cellular component synthesis and metabolic and biosynthetic processes. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis suggested that the plant hormone signal transduction pathway was significantly enriched. The common DEGs and four senescence-related pathways, including plant hormone signal transduction, porphyrin and chlorophyll metabolism, carotenoid biosynthesis, and regulation of autophagy were selected to be discussed further. This work aimed to provide potential genes signaling and modulating premature senescence as well as the possible dynamic network of gene expression patterns for further study.
Keywords: plant hormone; premature senescence; tobacco; transcriptome; ultrastructure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Transcriptome divergence between developmental senescence and premature senescence in Nicotiana tabacum L.Sci Rep. 2020 Nov 25;10(1):20556. doi: 10.1038/s41598-020-77395-2. Sci Rep. 2020. PMID: 33239739 Free PMC article.
-
Comparative transcriptomic analysis reveals that multiple hormone signal transduction and carbohydrate metabolic pathways are affected by Bacillus cereus in Nicotiana tabacum.Genomics. 2020 Nov;112(6):4254-4267. doi: 10.1016/j.ygeno.2020.07.022. Epub 2020 Jul 15. Genomics. 2020. PMID: 32679071
-
Transcriptome Analysis of a Premature Leaf Senescence Mutant of Common Wheat (Triticum aestivum L.).Int J Mol Sci. 2018 Mar 10;19(3):782. doi: 10.3390/ijms19030782. Int J Mol Sci. 2018. PMID: 29534430 Free PMC article.
-
The Role and Regulation of Autophagy and the Proteasome During Aging and Senescence in Plants.Genes (Basel). 2019 Apr 2;10(4):267. doi: 10.3390/genes10040267. Genes (Basel). 2019. PMID: 30987024 Free PMC article. Review.
-
Towards systems biological understanding of leaf senescence.Plant Mol Biol. 2013 Aug;82(6):519-28. doi: 10.1007/s11103-012-9974-2. Epub 2012 Oct 13. Plant Mol Biol. 2013. PMID: 23065109 Review.
Cited by
-
Whole-Transcriptome Analysis Reveals Autophagy Is Involved in Early Senescence of zj-es Mutant Rice.Front Plant Sci. 2022 Jun 3;13:899054. doi: 10.3389/fpls.2022.899054. eCollection 2022. Front Plant Sci. 2022. PMID: 35720578 Free PMC article.
-
Comparative transcriptome analysis reveals genes involved in trichome development and metabolism in tobacco.BMC Plant Biol. 2024 Jun 13;24(1):541. doi: 10.1186/s12870-024-05265-4. BMC Plant Biol. 2024. PMID: 38872084 Free PMC article.
-
Integrative Analyses of Biochemical Properties and Transcriptome Reveal the Dynamic Changes in Leaf Senescence of Tobacco (Nicotiana tabacum L.).Front Genet. 2021 Dec 22;12:790167. doi: 10.3389/fgene.2021.790167. eCollection 2021. Front Genet. 2021. PMID: 35003224 Free PMC article.
-
Transcriptome divergence between developmental senescence and premature senescence in Nicotiana tabacum L.Sci Rep. 2020 Nov 25;10(1):20556. doi: 10.1038/s41598-020-77395-2. Sci Rep. 2020. PMID: 33239739 Free PMC article.
-
Metatranscriptomic Analysis of Multiple Environmental Stresses Identifies RAP2.4 Gene Associated with Arabidopsis Immunity to Botrytis cinerea.Sci Rep. 2019 Nov 18;9(1):17010. doi: 10.1038/s41598-019-53694-1. Sci Rep. 2019. PMID: 31740741 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

