Anaplastic Lymphoma Kinase (ALK) Receptor Tyrosine Kinase: A Catalytic Receptor with Many Faces
- PMID: 30400214
- PMCID: PMC6274813
- DOI: 10.3390/ijms19113448
Anaplastic Lymphoma Kinase (ALK) Receptor Tyrosine Kinase: A Catalytic Receptor with Many Faces
Abstract
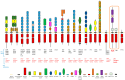
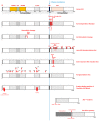
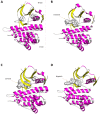
The anaplastic lymphoma kinase (ALK) receptor is a membrane-bound tyrosine kinase. The pathogenesis of several cancers is closely related to aberrant forms of ALK or aberrant ALK expression, including ALK fusion proteins, ALK-activated point mutations, and ALK amplification. Clinical applications of different ALK inhibitors represent significant progress in targeted therapy. Knowledge of different aspects of ALK biology can provide significant information to further the understanding of this receptor tyrosine kinase. In this mini-review, we briefly summarize different features of ALK. We also summarize some recent research advances on ALK fusion proteins in cancers.
Keywords: ALK; ALK fusion proteins; ALK kinase inhibitors; aberrant forms; cancers; neuroblastoma; targeted therapy.
Conflict of interest statement
The author declares no conflicts of interest.
Figures

References
-
- Shiota M., Fujimoto J., Semba T., Satoh H., Yamamoto T., Mori S. Hyperphosphorylation of a novel 80 kDa protein-tyrosine kinase similar to Ltk in a human Ki-1 lymphoma cell line, AMS3. Oncogene. 1994;9:1567–1574. - PubMed
-
- Morris S.W., Naeve C., Mathew P., James P.L., Kirstein M.N., Cui X., Witte D.P. ALK, the chromosome 2 gene locus altered by the t(2;5) in non-Hodgkin’s lymphoma, encodes a novel neural receptor tyrosine kinase that is highly related to leukocyte tyrosine kinase (LTK) Oncogene. 1997;14:2175–2188. doi: 10.1038/sj.onc.1201062. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

