G-Protein Coupled Receptor Protein Synthesis on a Lipid Bilayer Using a Reconstituted Cell-Free Protein Synthesis System
- PMID: 30400226
- PMCID: PMC6316570
- DOI: 10.3390/life8040054
G-Protein Coupled Receptor Protein Synthesis on a Lipid Bilayer Using a Reconstituted Cell-Free Protein Synthesis System
Abstract
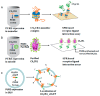
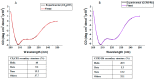
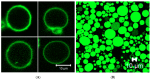
Membrane proteins are important drug targets which play a pivotal role in various cellular activities. However, unlike cytosolic proteins, most of them are difficult-to-express proteins. In this study, to synthesize and produce sufficient quantities of membrane proteins for functional and structural analysis, we used a bottom-up approach in a reconstituted cell-free synthesis system, the PURE system, supplemented with artificial lipid mimetics or micelles. Membrane proteins were synthesized by the cell-free system and integrated into lipid bilayers co-translationally. Membrane proteins such as the G-protein coupled receptors were expressed in the PURE system and a productivity ranging from 0.04 to 0.1 mg per mL of reaction was achieved with a correct secondary structure as predicted by circular dichroism spectrum. In addition, a ligand binding constant of 27.8 nM in lipid nanodisc and 39.4 nM in micelle was obtained by surface plasmon resonance and the membrane protein localization was confirmed by confocal microscopy in giant unilamellar vesicles. We found that our method is a promising approach to study the different classes of membrane proteins in their native-like artificial lipid bilayer environment for functional and structural studies.
Keywords: G-protein coupled receptor; artificial cell; cell-free protein synthesis; lipid bilayer; lipid nanodisc; membrane protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

