Circular RNAs in Muscle Function and Disease
- PMID: 30400273
- PMCID: PMC6274904
- DOI: 10.3390/ijms19113454
Circular RNAs in Muscle Function and Disease
Abstract
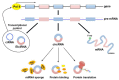
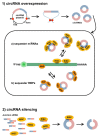
Circular RNAs (circRNAs) are a class of RNA produced during pre-mRNA splicing that are emerging as new members of the gene regulatory network. In addition to being spliced in a linear fashion, exons of pre-mRNAs can be circularized by use of the 3' acceptor splice site of upstream exons, leading to the formation of circular RNA species. In this way, genetic information can be re-organized, increasing gene expression potential. Expression of circRNAs is developmentally regulated, tissue and cell-type specific, and shared across eukaryotes. The importance of circRNAs in gene regulation is now beginning to be recognized and some putative functions have been assigned to them, such as the sequestration of microRNAs or proteins, the modulation of transcription, the interference with splicing, and translation of small proteins. In accordance with an important role in normal cell biology, circRNA deregulation has been reported to be associated with diseases. Recent evidence demonstrated that circRNAs are highly expressed in striated muscle tissue, both skeletal and cardiac, that is also one of the body tissue showing the highest levels of alternative splicing. Moreover, initial studies revealed altered circRNA expression in diseases involving striated muscle, suggesting important functions of these molecules in the pathogenetic mechanisms of both heart and skeletal muscle diseases. The recent findings in this field will be described and discussed.
Keywords: cardiac muscle; circular RNAs; muscle disease; skeletal muscle.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Edmondson D.G., Lyons G.E., Martin J.F., Olson E.N. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development. 1994;120:1251–1263. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

