SIFR annotator: ontology-based semantic annotation of French biomedical text and clinical notes
- PMID: 30400805
- PMCID: PMC6218966
- DOI: 10.1186/s12859-018-2429-2
SIFR annotator: ontology-based semantic annotation of French biomedical text and clinical notes
Abstract
Background: Despite a wide adoption of English in science, a significant amount of biomedical data are produced in other languages, such as French. Yet a majority of natural language processing or semantic tools as well as domain terminologies or ontologies are only available in English, and cannot be readily applied to other languages, due to fundamental linguistic differences. However, semantic resources are required to design semantic indexes and transform biomedical (text)data into knowledge for better information mining and retrieval.
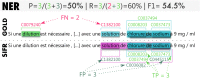
Results: We present the SIFR Annotator ( http://bioportal.lirmm.fr/annotator ), a publicly accessible ontology-based annotation web service to process biomedical text data in French. The service, developed during the Semantic Indexing of French Biomedical Data Resources (2013-2019) project is included in the SIFR BioPortal, an open platform to host French biomedical ontologies and terminologies based on the technology developed by the US National Center for Biomedical Ontology. The portal facilitates use and fostering of ontologies by offering a set of services -search, mappings, metadata, versioning, visualization, recommendation- including for annotation purposes. We introduce the adaptations and improvements made in applying the technology to French as well as a number of language independent additional features -implemented by means of a proxy architecture- in particular annotation scoring and clinical context detection. We evaluate the performance of the SIFR Annotator on different biomedical data, using available French corpora -Quaero (titles from French MEDLINE abstracts and EMEA drug labels) and CépiDC (ICD-10 coding of death certificates)- and discuss our results with respect to the CLEF eHealth information extraction tasks.
Conclusions: We show the web service performs comparably to other knowledge-based annotation approaches in recognizing entities in biomedical text and reach state-of-the-art levels in clinical context detection (negation, experiencer, temporality). Additionally, the SIFR Annotator is the first openly web accessible tool to annotate and contextualize French biomedical text with ontology concepts leveraging a dictionary currently made of 28 terminologies and ontologies and 333 K concepts. The code is openly available, and we also provide a Docker packaging for easy local deployment to process sensitive (e.g., clinical) data in-house ( https://github.com/sifrproject ).
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
CISMeF/SIBM (see related work section) were an early partner of the SIFR project.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

