Genome sequence of the potato pathogenic fungus Alternaria solani HWC-168 reveals clues for its conidiation and virulence
- PMID: 30400851
- PMCID: PMC6219093
- DOI: 10.1186/s12866-018-1324-3
Genome sequence of the potato pathogenic fungus Alternaria solani HWC-168 reveals clues for its conidiation and virulence
Abstract
Background: Alternaria solani is a known air-born deuteromycete fungus with a polycyclic life cycle and is the causal agent of early blight that causes significant yield losses of potato worldwide. However, the molecular mechanisms underlying the conidiation and pathogenicity remain largely unknown.
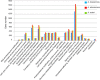
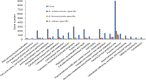
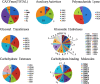
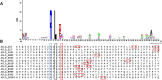
Results: We produced a high-quality genome assembly of A. solani HWC-168 that was isolated from a major potato-producing region of Northern China, which facilitated a comprehensive gene annotation, the accurate prediction of genes encoding secreted proteins and identification of conidiation-related genes. The assembled genome of A. solani HWC-168 has a genome size 32.8 Mb and encodes 10,358 predicted genes that are highly similar with related Alternaria species including Alternaria arborescens and Alternaria brassicicola. We identified conidiation-related genes in the genome of A. solani HWC-168 by searching for sporulation-related homologues identified from Aspergillus nidulans. A total of 975 secreted protein-encoding genes, which might act as virulence factors, were identified in the genome of A. solani HWC-168. The predicted secretome of A. solani HWC-168 possesses 261 carbohydrate-active enzymes (CAZy), 119 proteins containing RxLx[EDQ] motif and 27 secreted proteins unique to A. solani.
Conclusions: Our findings will facilitate the identification of conidiation- and virulence-related genes in the genome of A. solani. This will permit new insights into understanding the molecular mechanisms underlying the A. solani-potato pathosystem and will add value to the global fungal genome database.
Keywords: Alternaria solani; Conidiation; Genome sequence; Secretome; Virulence.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
Transcriptome sequencing leads to an improved understanding of the infection mechanism of Alternaria solani in potato.BMC Plant Biol. 2023 Mar 1;23(1):120. doi: 10.1186/s12870-023-04103-3. BMC Plant Biol. 2023. PMID: 36859112 Free PMC article.
-
Gapless Genome Assembly of the Potato and Tomato Early Blight Pathogen Alternaria solani.Mol Plant Microbe Interact. 2018 Jul;31(7):692-694. doi: 10.1094/MPMI-12-17-0309-A. Epub 2018 May 18. Mol Plant Microbe Interact. 2018. PMID: 29432053
-
Alternaria solani effectors AsCEP19 and AsCEP20 reveal novel functions in pathogenicity and conidiogenesis.Microbiol Spectr. 2024 Aug 6;12(8):e0421423. doi: 10.1128/spectrum.04214-23. Epub 2024 Jun 24. Microbiol Spectr. 2024. PMID: 38912810 Free PMC article.
-
How the necrotrophic fungus Alternaria brassicicola kills plant cells remains an enigma.Eukaryot Cell. 2015 Apr;14(4):335-44. doi: 10.1128/EC.00226-14. Epub 2015 Feb 13. Eukaryot Cell. 2015. PMID: 25681268 Free PMC article. Review.
-
Alternaria diseases on potato and tomato.Mol Plant Pathol. 2024 Mar;25(3):e13435. doi: 10.1111/mpp.13435. Mol Plant Pathol. 2024. PMID: 38476108 Free PMC article. Review.
Cited by
-
Genome assembly and comparative analysis of Alternaria Linariae reveal novel genes associated with host colonization and virulence.BMC Genomics. 2025 Jul 7;26(1):638. doi: 10.1186/s12864-025-11819-z. BMC Genomics. 2025. PMID: 40624476 Free PMC article.
-
Transcriptome sequencing leads to an improved understanding of the infection mechanism of Alternaria solani in potato.BMC Plant Biol. 2023 Mar 1;23(1):120. doi: 10.1186/s12870-023-04103-3. BMC Plant Biol. 2023. PMID: 36859112 Free PMC article.
-
Antifungal Effects of Volatiles Produced by Bacillus subtilis Against Alternaria solani in Potato.Front Microbiol. 2020 Jun 17;11:1196. doi: 10.3389/fmicb.2020.01196. eCollection 2020. Front Microbiol. 2020. PMID: 32625175 Free PMC article.
-
Multi-omics approaches to understand pathogenicity during potato early blight disease caused by Alternaria solani.Front Microbiol. 2024 Mar 11;15:1357579. doi: 10.3389/fmicb.2024.1357579. eCollection 2024. Front Microbiol. 2024. PMID: 38529180 Free PMC article.
-
Bacillus velezensis Strain HN-Q-8 Induced Resistance to Alternaria solani and Stimulated Growth of Potato Plant.Biology (Basel). 2023 Jun 14;12(6):856. doi: 10.3390/biology12060856. Biology (Basel). 2023. PMID: 37372140 Free PMC article.
References
-
- GamboaAngulo MM, AlejosGonzalez F, PenaRodriguez LM. Homozinniol, a new phytotoxic metabolite from Alternaria solani. J Agric Food Chem. 1997;45(1):282–285. doi: 10.1021/jf960134p. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

